Editor’s note: The researchers would like to acknowledge co-authors Christopher Takahashi, Gagan Gupta, Jake Smith, Richard Rouse, Paul Berndt, Sergey Yekhanin, David Ward, Siena Ang, Patrick Garvan, Hsing-Yeh Parker, Rob Carlson, Douglas Carmean, and Luis Ceze for their contributions to this work.
Current estimates by the International Data Corporation indicate a 20.4 percent year-over-year growth in demand for data storage, which is predicted to reach almost 9 zettabytes by 2024 (opens in new tab). To put that number into perspective, Windows 11, which takes up around 64 gigabytes (opens in new tab) of storage space initially, would need to be installed on over 15 billion devices to add up to just 1 zettabyte of storage used. For comparison, just over 3 billion personal computers are estimated to have shipped worldwide since 2011 (opens in new tab).
Available methods of storage are having difficulty keeping up with the increasing demand in the long term. Synthetic DNA, at its root a tiny storer of information, offers a potential pathway toward significantly reducing the amount of space and material needed for future archival storage needs. Revisiting the projection of growth above, it would take millions of tape cartridges—the current densest commercial storage media—to store 9 zettabytes of information, whereas it would take the footprint of one small refrigerator if stored in DNA.
Spotlight: blog post
DNA not only offers a clear advantage over existing storage media with density at over 1 exabyte per cubic inch, but also is a potential solution to present-day challenges in data archival storage systems. DNA can be incredibly durable and can last thousands of years, unlike tape, which needs to be rewritten every 30 years at best. DNA data storage will not go obsolete since the techniques to read the DNA molecules are numerous and relevant to life science applications. Additionally, evidence points to potential for DNA storage to have lower greenhouse gas emissions, water consumption, and energy consumption. Despite these advantages, one key impediment to large-scale deployment of DNA data storage has been its low DNA synthesis throughput, resulting in low throughput of writing and relatively high cost.

We have been working with our University of Washington collaborators at the Molecular Information Systems Laboratory (MISL) to address this problem. Our paper published in Science Advances, “Scaling DNA Data Storage with Nanoscale Electrode Wells,” introduces a proof-of-concept molecular controller in the form of a tiny DNA storage writing mechanism on a chip. The chip demonstrates the ability to pack DNA-synthesis spots three orders of magnitude more tightly than before. This shows that much higher DNA writing throughput can be achieved.
In this blog post, we’ll discuss the write/read processes of DNA storage, the advances our work makes to show writing throughput can increase for more widespread storage needs, and the technology we created to attain this achievement, including a nanoscale electrochemical array. Ultimately, we were able to use the system to encode a message onto four strands of synthetic DNA, proof that nanoscale DNA writing is possible at dimensions necessary for practical DNA data storage.
Writing and reading synthetic DNA means translating bits to molecules and back again
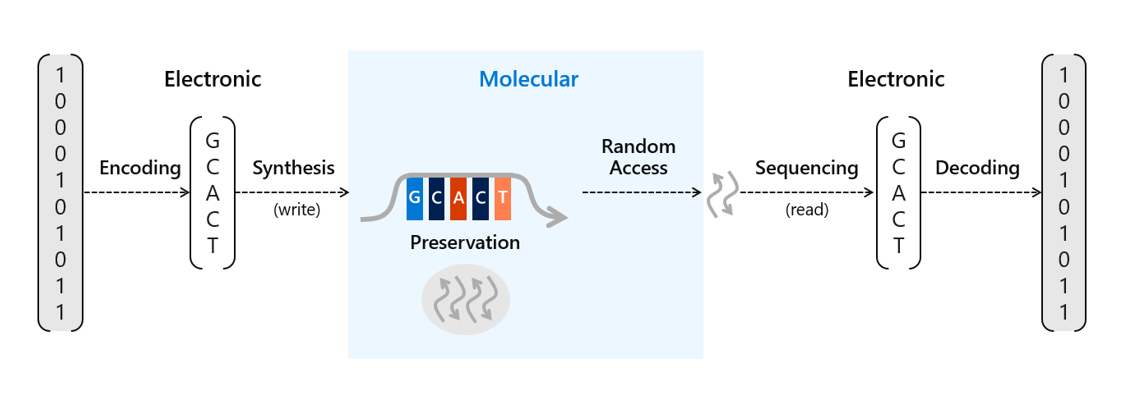
There’s been a lot of work done recently to advance the potential scale of DNA storage, such as developing automation techniques to avoid the laborious process of manually pipetting DNA and other reagents or methods to protect the DNA for long-term storage in excess of thousands of years. We’ve already conducted research into a number of areas of DNA data storage, including an end-to-end data storage system capable of random access and viable methods for preservation of DNA. Storing information in DNA at the scale necessary for commercial use requires two crucial processes. The first requires translating digital bits (ones and zeros) into strands of synthetic DNA representing these bits with encoding software and a DNA synthesizer. The second is to read and decode the information back into bits to recover that information into digital form again with a DNA sequencer and decoding software.
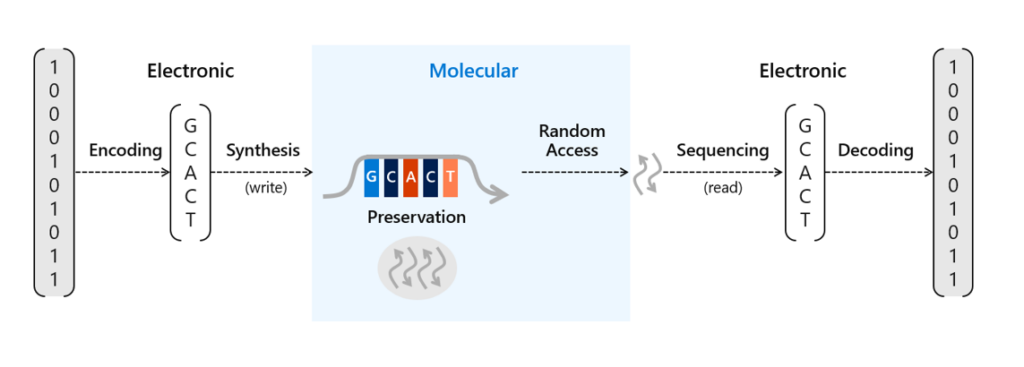
To store data on synthetic DNA, digital bits are encoded in the DNA bases (A, C, T, and G) of a DNA sequence. When data is stored in DNA, a DNA chain containing the specific sequence of bases must be synthesized (see Figure 1). DNA chains are traditionally created through a multi-step technique called phosphoramidite chemistry. In this process, a DNA chain is grown by the sequential addition of the DNA bases. Each DNA base contains a blocking group that prevents multiple additions of the base to the growing DNA chain. Once a blocked base is attached to the DNA chain, acid is delivered to cleave the blocking group and prime the DNA chain for the addition of the next base.
Synthesizing DNA chains can be done individually or in parallel on an array, which contains multiple spots where unique DNA sequences can be created simultaneously. Increasing synthesis density, that is, the number of synthesis spots on a fixed area, is the key to increasing the writing throughput and lowering its cost. The closer together these spots are on an array, the lower the synthesis cost of each DNA chain because the materials needed for the process can be used with more sequences.
Electrochemical arrays open the door to nanoscale feature sizes
The main challenge to increasing DNA writing throughput is to maintain control of individual spots without interfering with neighboring spots. Current DNA synthesis arrays are designed for generating a small number of high-quality DNA sequences with millions of exact copies and are achieved through three main array synthesis methods: photochemistry, fluid deposition, and electrochemistry.
In photochemical DNA synthesis, a photomask or micromirror creates patterns of light on an array, which removes the blocking group from the DNA strand. Liquid deposition, such as acoustic or inkjet printing methods, deliver the acid deblock to the individual spots. Both methods, however, are limited in the synthesis densities they can attain due to micromirror size, light scattering, or droplet stability. Electrochemical arrays, however, can leverage the semiconductor roadmap where 7 nanometer (nm) feature sizes are common.
In electrochemical DNA synthesis, each spot in the array contains an electrode. Once a voltage is applied, acid is generated at the anode (working electrode) to deblock the growing DNA chains, and an equivalent base is generated at the cathode (counter electrode). The main concern when scaling down the pitch between anodes is acid diffusion; the smaller the pitch, the easier it may be for acid to diffuse to neighboring electrodes and cause unintended deblocking at those locations. While commercial electrochemical arrays have demonstrated acid generation and minimal diffusion at micron-sized electrodes, it was not clear if this trend would continue indefinitely to smaller features.
Our colleagues at MISL modeled acid generation at small feature sizes using finite elemental analysis to determine whether this trend would hold. We adopted a design layout containing a 650-nm electrode embedded within a glass well surrounded by cathodes. The glass well would serve as the attachment surface to grow the DNA chains and as a physical barrier to prevent acid from diffusing to neighboring sites. As an extra buffer, any acid that escaped the well would encounter base generated at the cathodes and be neutralized. The model suggested that acid could be confined at these and even smaller scales, encouraging us to design chips containing small feature spots and to get them manufactured.

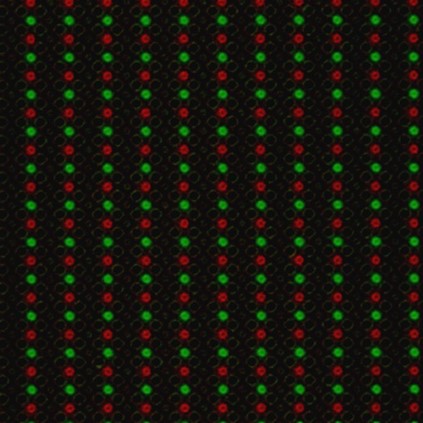
To put theory into practice, we had chip arrays fabricated with the previously mentioned layout. These electrochemical arrays contained sets of four individually addressable electrodes. With them, we demonstrated the ability to control DNA synthesis at desired locations by performing experiments with two fluorescently labeled bases (green and red). If acid were diffusing unexpectedly, it would reach unintended spots and we would see one color bleed over other spots.
On an electrochemical array, we generated acid at one set of electrodes to deblock the DNA chain and then added a green-fluorescent base. In the next step, we generated acid at a different set of electrodes of the same array and coupled a red-fluorescent base to generate the image seen in Figure 3. As expected, we saw no bleed over, confirming we had no unintended acid diffusion.
On a separate array, we then demonstrated the array’s capability to write data by synthesizing four unique DNA strands, each 100 bases long, which encoded the motto “Empowering each person to store more!” Although the error rates were higher than commercial DNA synthesizers, we could still decode the message with no bit errors.
The path forward for nanoscale molecular controllers
Our proof of concept paves a road toward generating massive numbers of unique DNA sequences in parallel for data storage. By injecting electrons at specific locations, we can control the molecular environment surrounding the electrodes and thus control the sequence of the DNA grown there. A natural next step is to embed digital logic in the chip to allow individual control of millions of electrode spots to write kilobytes per second of data in DNA. From there, we foresee the technology reaching arrays containing billions of electrodes capable of storing megabytes per second of data in DNA. This will bring DNA data storage performance and cost significantly closer to tape. We welcome further discussions to fully realize more widely available molecular controllers in the future.
Our work used phosphoramidite chemistry, which is standard today in biotechnology. However, this chemistry is based on a fossil-based solvent. To deliver on DNA’s sustainability promise, we expect water-based solutions to replace fossil-based chemistries. We have been evaluating new enzymatic DNA synthesis platforms, such as DNA Script’s SYNTAX System. We’re also working with Ansa Biotechnologies to develop a set of enzymatic reagents that are specifically tailored for our electrochemical array. We are always interested in learning about new chemistries and other technology that could make DNA data storage more sustainable.
More broadly, this work demonstrates control over the electronic-to-molecular interface, which we posit opens a door to new applications. For example, electrochemical control methods enable spatial control of enzymes at the nanoscale. Beyond DNA, this could also be a tool for drug discovery, by enabling rapid combinatorial organic synthesis as a platform for screening drug-protein binding kinetics. Other examples are a tool for assays that detect disease biomarkers or even a platform for sensing environmental pollutants.
If you’d like to learn more about the DNA storage work happening at Microsoft Research, check out the DNA Storage project page (opens in new tab). You can also further explore how electronic-molecular systems store data, and can perform certain types of computation, in a Microsoft Research webinar with Karin Strauss (opens in new tab). To learn more about the emerging industry of DNA data storage, check out the DNA Data Storage Alliance (opens in new tab) page.