
Microsoft Research and Cyted have collaborated to build novel AI models (opens in new tab) to scale the early detection of esophageal cancer. The AI-supported methods demonstrated the same diagnostic performance as the existing manual workflow, potentially reducing the pathologist’s workload by up to 63%.
Esophageal cancer is the sixth most common cause of cancer deaths worldwide, in part because this disease is typically diagnosed late, making treatment difficult. Fewer than 1 in 5 patients survive five years after diagnosis, making early detection of this disease critical to improving a patient’s chances. One opportunity for early detection is to identify patients with a condition called Barrett’s esophagus (BE). Patients with BE are at an increased risk of developing cancer, though most never will. Chronic heartburn is a risk factor and a possible cause of Barrett’s.
Detecting BE dramatically improves a patient’s chances. Earlier detection of cancer and earlier start of treatment mean that more than 9 in 10 patients survive 5 years after diagnosis. However early detection of BE has typically involved an endoscopic biopsy, a procedure that many people find uncomfortable and invasive. It often requires sedation, is resource intensive, and increases the risk of complications.
Spotlight: Microsoft research newsletter
A major step toward enabling large-scale screening for BE has been spearheaded by Cyted (opens in new tab), a start-up company at the forefront of medical innovation. Cyted has developed a capsule sponge device called EndoSign (opens in new tab)® – a dissolvable capsule on a string that expands into a small medical sponge once in the stomach. When pulled back out, it collects cells from the lining of the esophagus, which are then processed, placed on slides, stained, and scanned for digital analysis.
The capsule sponge is easier to administer and less costly than endoscopy. But a pathologist still needs to review the digitized slides to determine the presence of any goblet cells, a type of cell normally found in the intestinal lining, which would indicate BE if found in the esophagus. These images are huge (up to 100,000 by 100,000 pixels – the size of a squash court if printed at the typical photo resolution of 300dpi) – yet may contain only a few goblet cells per image, each cell just a few pixels large. To identify BE, pathologists need to use slides from two stains, H&E (a routine stain for observing cell structure) and TFF3 (a special stain just to find goblet cells). Since most patients with heartburn will not have BE, pathologists spend most of their time examining negative cases, taking away time in which they could be prioritizing high-risk cases without more sophisticated approaches to analysis.
Microsoft Research and Cyted have collaborated to build novel AI models that can efficiently check the slides for goblet cells, using either the H&E or TFF3 stains. This joint effort has led to a Nature Communications paper titled “Enabling large-scale screening of Barrett’s esophagus using weakly supervised deep learning in histopathology (opens in new tab).” Our study uses the strength of transformer-based multiple instance learning to assist in the screening of BE. In the paper, we introduce two major innovations. First, we show that the AI models can be built solely from the pathologists’ findings on whether BE is present, eliminating the need for expensive pixel-level annotations. This means that existing large capsule sponge screening datasets can be used to further improve the performance of the model. Secondly, we demonstrate that goblet cells can be detected with high accuracy using only the H&E slides. This is the most common routine stain in pathology, and it suggests that the more time-consuming and costly specialized staining, TFF3, could be skipped (see Figure 2 below).
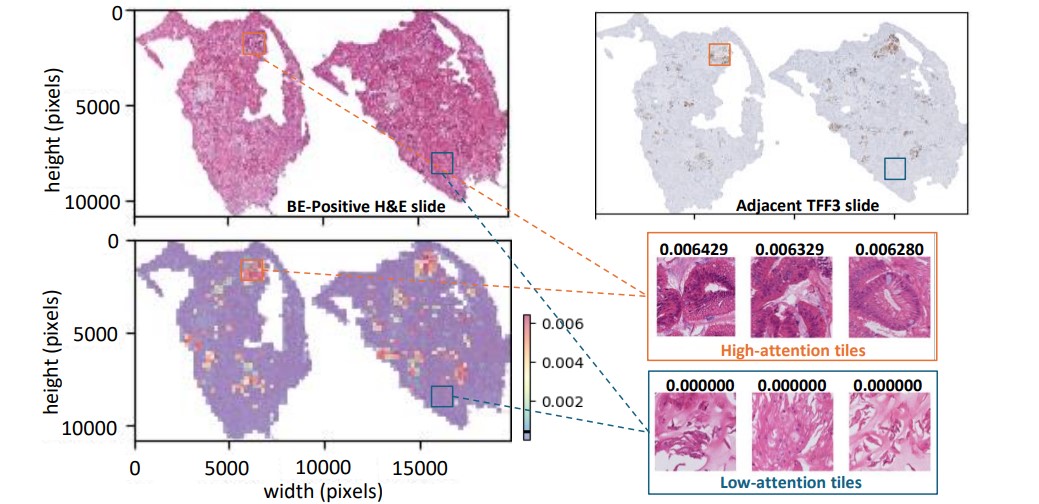
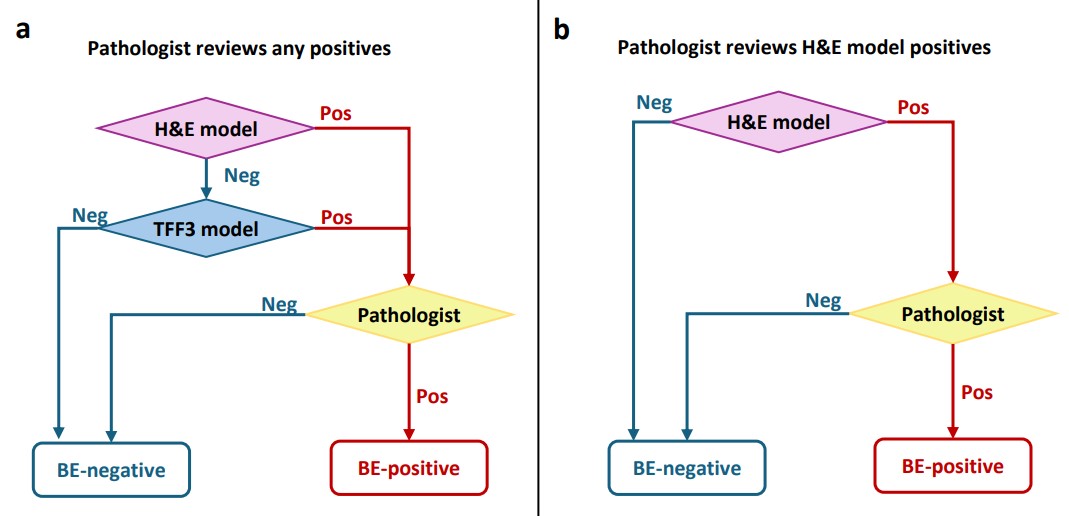
In the paper, we further discuss different AI-assisted workflows designed to optimize the screening process. The first workflow necessitates a pathologist’s review only if either the H&E or TFF3 models predict a sample as positive. This method can achieve the same diagnostic performance as the existing manual workflow in terms of sensitivity and specificity, potentially reducing the pathologist’s workload by 52% (see Figure 3 below).
The second proposed workflow reduces the need for pathologist review by 63% of the original load, by restricting reviews to positive predictions from the H&E model only. However, this comes at slightly reduced sensitivity, since goblet cells are more clearly visible in the TFF3 stain.
| Proposed AI-assisted workflow | Pathologist review (per-cent of all cases) | TFF3 staining required (per-cent of all cases) | Sensitivity @ Specificity 1.00 |
|---|---|---|---|
| Pathologist reviews any positive | 48% | 100% | 1.00 |
| Pathologist reviews H&E model positives | 37% | 37% | 0.91 |
Our collaboration with Cyted demonstrates the transformative potential of integrating advanced AI models into clinical workflows, saving valuable time for pathologists. As we move forward, the scalability of this technology holds the promise for widespread adoption of early detection in the fight against esophageal cancer.
“This represents a significant step in our fight against esophageal cancer, offering the potential to save countless lives through early detection with our minimally-invasive capsule sponge technology,” said Cyted CEO Marcel Gehrung. “Our collaboration with Microsoft Research has been instrumental in pushing the boundaries of what’s possible in medical imaging and screening technologies, creating optimal efficiencies from start to finish of the testing process.”
We have open sourced code to build these models (opens in new tab), which is designed to be scalable to very large datasets, using Azure Machine Learning (opens in new tab). This flexibility allows other researchers and institutions to adapt and enhance our code according to their specific needs. Importantly, our code represents a significant advancement over previous work in the field. Unlike earlier approaches that focused solely on training the multiple instance and attention layers, our code allows for end-to-end fine-tuning, including the image encoder. This comprehensive approach to training ensures optimal performance and accuracy, setting a new standard for AI models in histopathology.
“The open sourcing of this code has helped us to advance our research in the field of early cancer detection,” said Florian Markowetz, Professor of Computational Oncology at the University of Cambridge, and Senior Group Leader at Cancer Research UK Cambridge Institute. “Several key features will soon be integrated into ongoing clinical trials, where we aim to improve the detection of Barrett’s esophagus in patients and ultimately treat more cancers through early intervention. Furthermore, these features will help improve the workflow of pathologists and identify key regions quicker, enabling clinicians to tackle more cases with greater reliability.”
By sharing our work, we aim not only to enhance the detection of BE and esophageal cancer, but also to empower researchers and clinicians around the world to leverage this technology in their fight against cancer[1]. Because our code can be used as a building block to develop AI models for histopathology slides, it may also potentially be applied to other cancer types. It is our hope that this open-source initiative will foster innovation and collaboration, and ultimately lead to breakthroughs that save lives.
As researchers, it has been exciting to work closely with Cyted and be part of the long path towards early detection of esophageal cancer. Cross-discipline collaborations like this are excellent opportunities to solve complex clinical problems. With AI models built using the principles of responsible AI like fairness, privacy and security, and reliability and safety, we can ultimately make a tangible difference to patient outcomes.
Acknowledgement
Thank you to the team: Kenza Bouzid, Harshita Sharma, Sarah Killcoyne, Daniel C. Castro, Anton Schwaighofer, Max Ilse, Valentina Salvatelli, Ozan Oktay, Sumanth Murthy, Lucas Bordeaux, Luiza Moore, Maria O’Donovan, Anja Thieme, Hannah Richardson, Aditya Nori, Marcel Gehrung, Javier Alvarez-Valle
[1] (opens in new tab) Code released for research use only. Full disclaimer here: https://github.com/microsoft/be-trans-mil (opens in new tab)





