All around us, biochemical systems are regulating a variety of natural processes, from our body’s ability to protect our skin through the timely production of melanin to plants’ ability to convert carbon dioxide into carbohydrates and oxygen using sunlight. Replicating and programming such complex systems—essentially creating computing networks capable of operating in biological environments—offers a unique opportunity to go where traditional silicon-based computers can’t. For example, with synthetic biocompatible controllers, we’re looking at the potential for targeted medical therapies. Think cancer treatments that attack only dangerous cells, sparing healthy ones, or capsules that delivery drugs or antibodies at opportune times.
Building these systems requires programming chemicals, and synthetic DNA is an ideal raw material to work with. Highly programmable, like transistors in silicon technology, and biocompatible, DNA can be used to implement chemical reaction networks (CRNs), a programming language for representing chemistry and biological processes, or the algorithmic and logical functions in traditional computing terms. Existing architectures implementing CRNs via DNA come in two varieties: DNA-only systems and multienzyme DNA systems. Researchers have been using DNA-only systems to build interesting digital logic circuits, such as circuits that can compute the square root of four-bit numbers (opens in new tab) and others that can classify handwritten digits (opens in new tab) by implementing pretrained neural networks. But both DNA-only and multienzyme DNA architectures have drawbacks, mainly slow rates and leaky reactions for DNA-only systems and increased biological complexity that can restrict environmental conditions for multienzyme systems. These challenges can limit the size of the systems, as well as their introduction into biological environments.
Spotlight: On-demand video
We propose a promising solution with a novel method for implementing chemical reaction networks that incorporates one enzyme. The idea is to replace the most fundamental unit of DNA computing, namely, toehold mediated strand displacement (TMSD), a DNA-only architecture, with polymerase-based strand displacement (PSD). Using DNA polymerase-based systems has several benefits: The polymerase enzyme gives an external energy source to the system, which is usually required; can synthesize new DNA strands, unlike enzyme-free systems; and can be potentially really fast. We present polymerase-based strand displacement in the paper “Using Strand Displacing Polymerase to Program Chemical Reaction Networks,” which was published in the Journal of the American Chemical Society.
Toehold mediated strand displacement vs. polymerase-based strand displacement
In 2006, researchers from the California Institute of Technology introduced enzyme-free logic computing (opens in new tab), which has since become the go-to approach for programming chemical reaction networks among computer scientists. Because of its simplicity and tunability, TMSD in particular has become one of the most fundamental DNA computing architectures. TMSD and PSD are similar in design, though. But first, a few things about DNA.
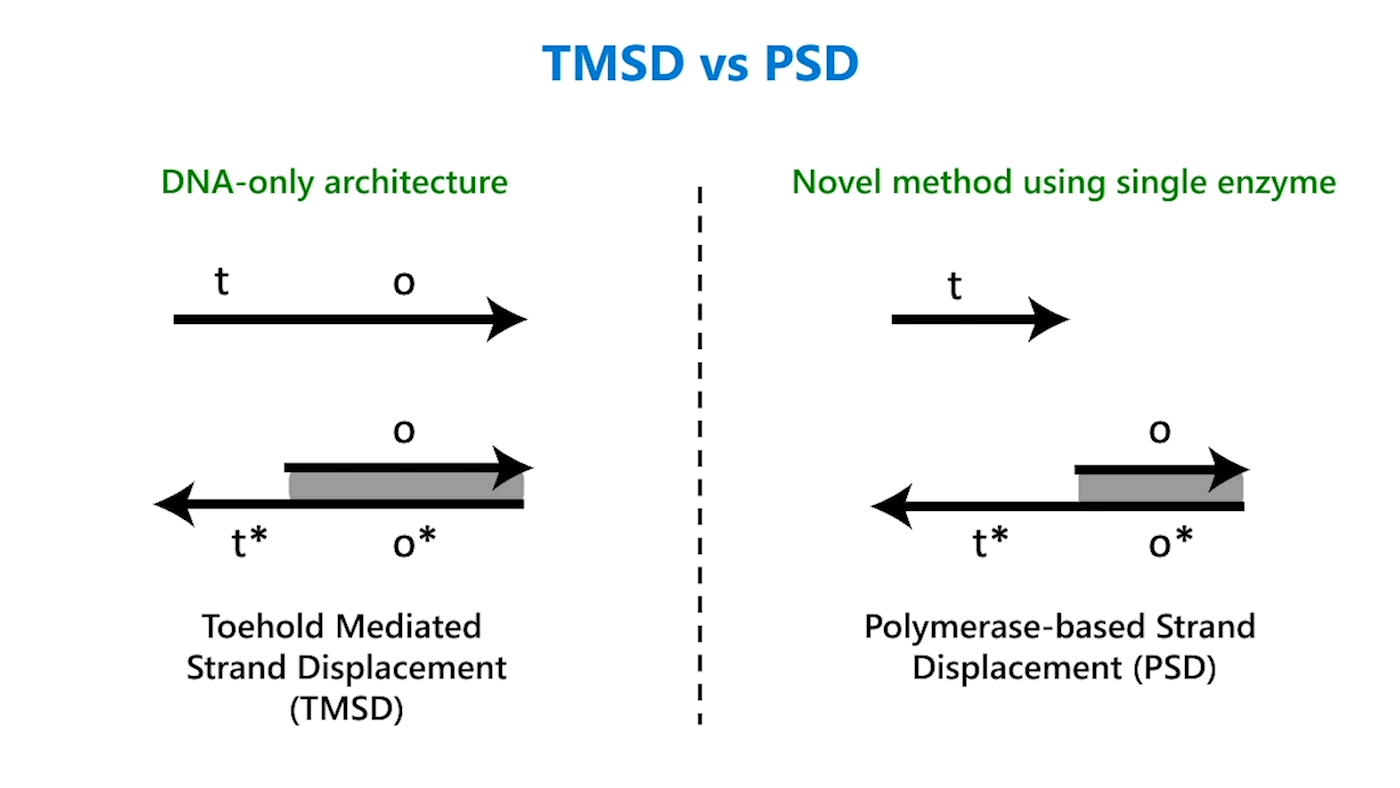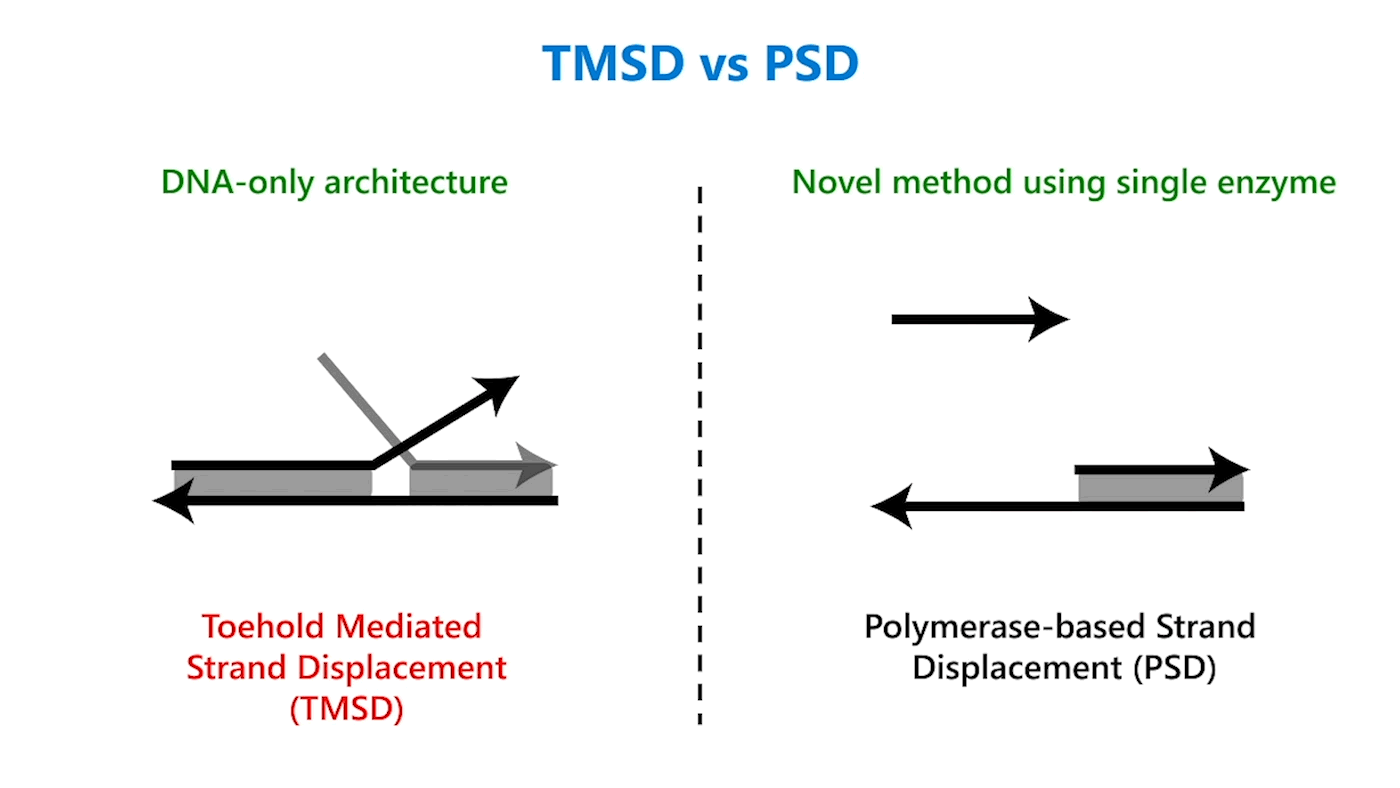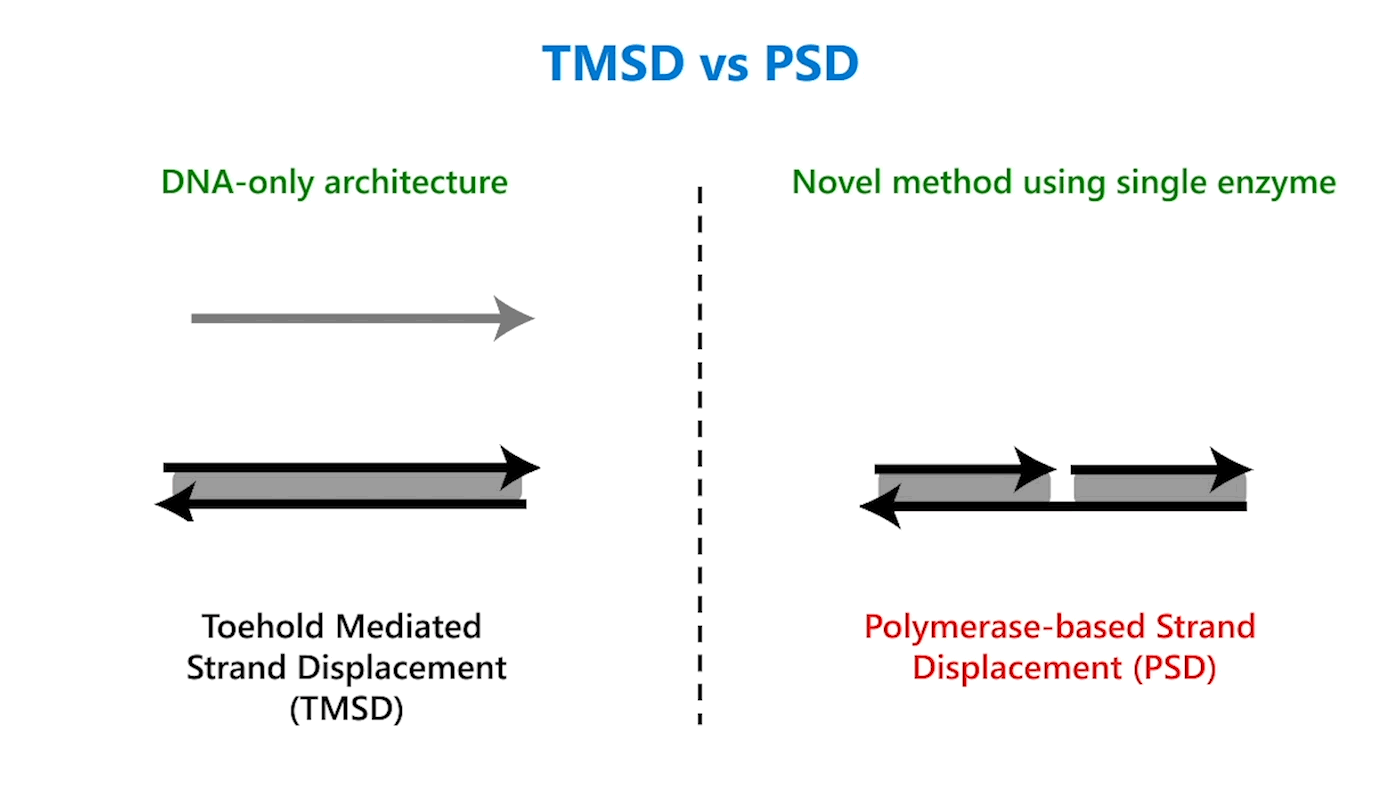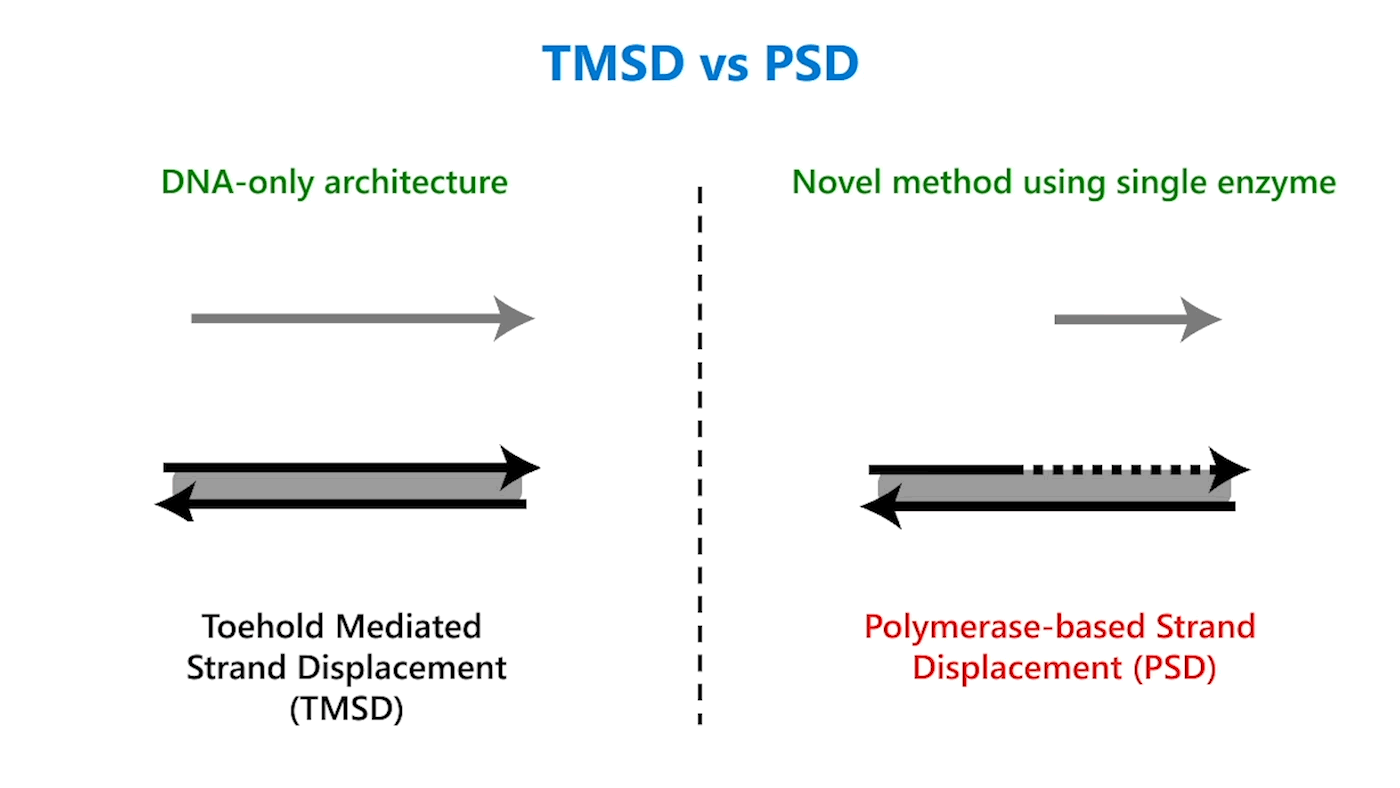
DNA strands are composed of sequences of the chemical bases adenine (A), guanine (G), cytosine (C), and thymine (T); DNA strands combine to form complexes, with A joining to its complementary base T and C to its complementary base G. Usually, an abstract representation of DNA strands is used to make it easier to design complicated architectures, which can have multiple DNA strands, each with sequences numbering in the tens or hundreds. So, for example, a portion of a single-stranded DNA with the sequence TACGTATGAATCAG might be referred to as domain o (Figure 1a). The reverse complement of that sequence would be ATGCATACTTAGTC, or more simply domain o*. This way, we don’t need to worry about actual sequences but can operate abstractly at the domain level. Sequences on a DNA strand can also be split into different parts, each with a separate domain name. In the domain o example, the sequence could be split after the first G: TACG TATGAATCAG. The first portion of the sequence could be domain t and the second domain o; the overall strand is t o.
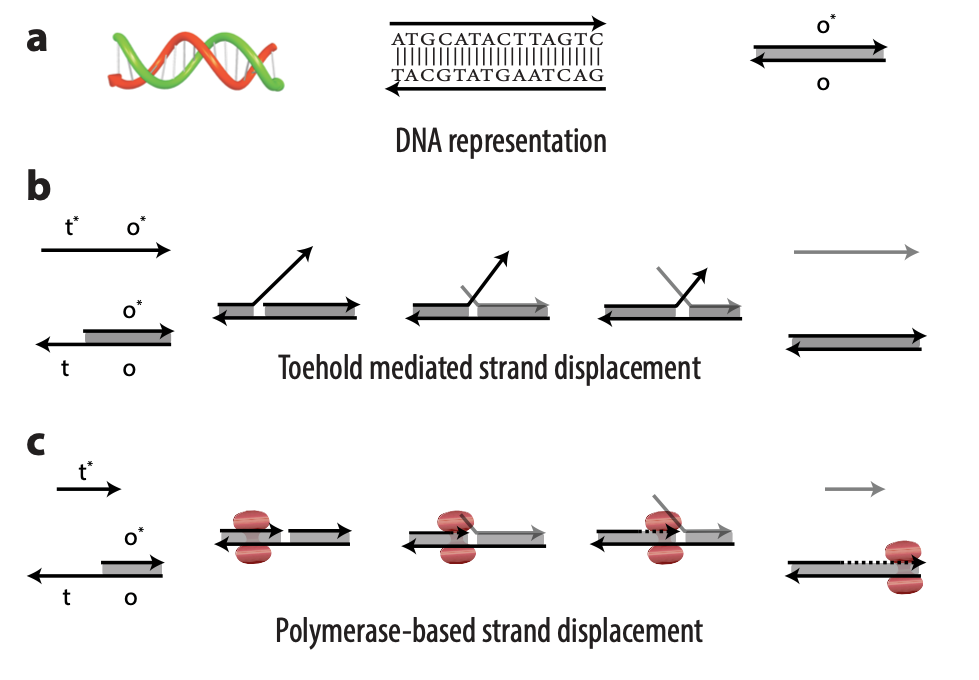
In TMSD, an input DNA strand consisting of complementary domains—let’s call the strand t* o*—binds with an exposed single-stranded, or toehold, portion of a double-stranded DNA complex t o (Figure 1b). This binding displaces the output strand in a “tug of war” between the duplicate portion of the input strand—the o* domain—and the o* domain that already exists in the complex. In PSD, a shorter input DNA strand without the o* domain attaches to the exposed portion of the complex; once there—and in our design, it’s a permanent bind—the polymerase enzyme elongates the input strand by printing new DNA, a t* o* copy, displacing the output in the double-stranded complex (Figure 1c).
Though similar in design, the addition of just one enzyme, polymerase, offers promising benefits. TMSD challenges include leaky reactions, which require workarounds that result in slower computation and/or greater system complexity. One reason for leaks is the presence of the output domain in the input, creating an overlap between the signals that increases the potential for error. In PSD, there is no overlap. The fact that there’s no longer a tug of war between strands, a time-consuming process, also contributes to faster computation times. Additionally, polymerase provides an external energy source, which allows for more complex computation within a reasonable time frame. For example, in 24 hours, DNA-only architectures may only be able to complete three layers of circuits; the faster computation offered by PSD can potentially complete 20 layers in the same period. This external energy source comes from the hydrolysis of the A, G, C, and T bases and the sugar and phosphate molecules that accompany them, collectively known as nucleoside triphosphates (NTPs). For example, it takes DNA-only architectures 10 hours to compute the square root function of a four-bit input; the faster computation offered by PSD can complete the same function within 40 minutes (opens in new tab). With PSD, since external NTPs and polymerase are used, several hundreds of bases can be displaced even if the toehold is short.
 (opens in new tab)
(opens in new tab)Designing unimolecular and bimolecular reactions
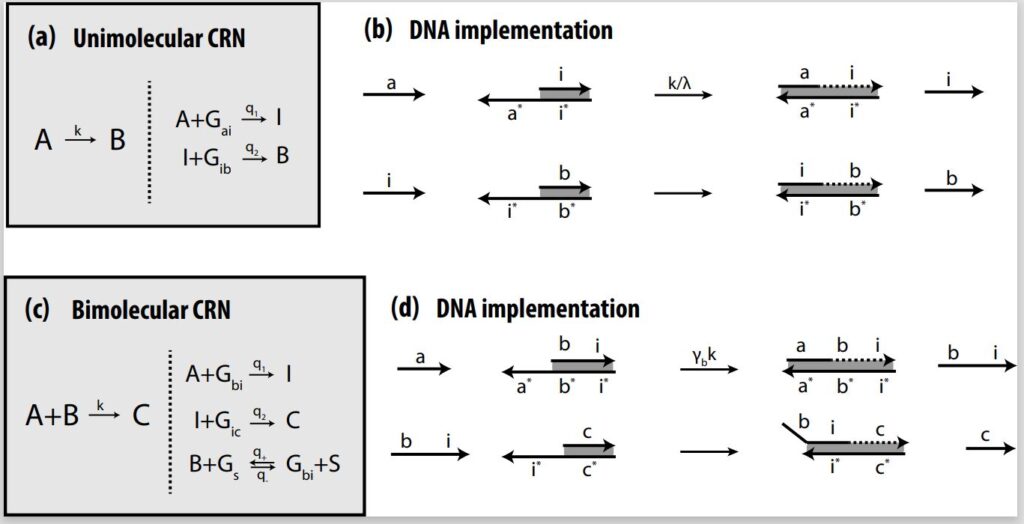
Much like logic gate AND and logic gate OR are considered universal gates in traditional computation, unimolecular and bimolecular CRNs are considered the two most fundamental gates in chemistry as far as design and implementation. In unimolecular reactions, only one reactant is used; in a bimolecular reaction, two reactants are used. Depending on the design, both CRNs are capable of producing either a single output or multiple outputs. Complex CRNs with more than two inputs and/or outputs can be built using these basic CRNs, enabling more advanced processes.
In theory, as described in the earlier paper “Implementing Arbitrary CRNs Using Strand Displacing Polymerase,” (opens in new tab) we demonstrate that we can implement unimolecular and bimolecular CRNs using PSD and do any arbitrary computations. We also apply these CRNs in an autocatalytic amplifier, a molecular-scale consensus network, and a dynamic rock-paper-scissor oscillatory system. In our architecture, unimolecular reactions and bimolecular reactions are both implemented in a two-step process. In the unimolecular reaction, A combines with an auxiliary gate to release intermediate strand I, which then combines with the next gate to produce output strand B. In the bimolecular reaction, input A combines with input B to produce intermediate strand I, which combines with the next gate to produce output strand C.
Implementing PSD in lab
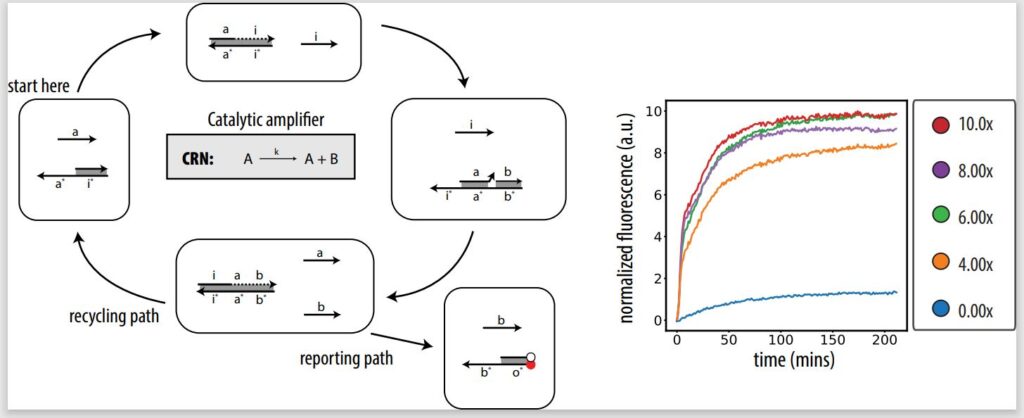
In our Journal of the American Chemical Society paper, we take the first step toward demonstrating in lab that our architecture can implement CRNs, showing the fundamentals of PSD systems and designing an in vitro catalytic amplifier. A catalytic amplifier is a unimolecular reaction with two outputs, but one of the outputs is also input. A catalytic amplifier is a strong proof of concept. Not only is it a show of complex DNA design, but it’s also sustainable.
Unlike in noncatalytic systems, where you need the same amount of input to trigger a certain amount of output, in a catalytic system like a catalytic amplifier, you can trigger 100 percent output with, let’s say, only 10 percent input. That’s because of the presence of a catalyst, or a fuel, which helps recycle the input so it can be used again to release more output. Even if there is a signal loss, you can still get experimentally full output activation from lower levels of input. For example, a catalytic amplifier could be used for fast signal restoration in deep circuits and neural nets.
In executing a catalytic amplifier, as well as other fundamental reactions, we show we’re able to tune two defining properties of a chemical reaction: stoichiometry and reaction rate. Our PSD architecture will only activate the same amount of output as input, allowing us to control the amount of output via the amount of input (stoichiometry), and the speed at which output is produced can be controlled by varying the lengths of the input strands.
Infinite possibilities
This was the first study on implementing CRNs using PSD, so for us, the goal was to explore the fundamentals. Our work mainly focuses on the design and demonstration of the basic properties of such systems, such as tuning the reaction speed and controlling the stoichiometry, and with the catalytic amplifier, we put the fundamentals to work. Next steps include in vitro demonstrations of larger-scale autocatalytic systems such as oscillators, linear controllers, and pulses, which are the accepted gold standards in the field. Such complex biochemical controllers can have computing applications in the biological context, where traditional silicon can’t reach, offering new possibilities in medical care, as mentioned above, agriculture, energy, molecular biology, computing, and sensor networks, among other areas.
DNA computing is decades behind its silicon counterpart, but we see fast growth as a real possibility, with the field learning from current technology. And thanks to design software like Visual DSD from Microsoft Research, DNA computing will be more streamlined, and designing and testing a new architecture—without leaks—should be easier, opening the way for some very cool applications.
Acknowledgment: This work was conducted by Shalin Shah, Jasmine Wee, Tianqi Song, Luis Ceze, and Karin Strauss, jointly led by John Reif at Duke University and Yuan-Jyue Chen at Microsoft Research. Shah was a Microsoft Research intern at the time of the work.