
Imagine if pathologists had tools that could help predict therapeutic responses just by analyzing images of cancer tissue. This vision may someday become a reality through the revolutionary field of computational pathology. By leveraging AI and machine learning, researchers are now able to analyze digitized tissue samples with unprecedented accuracy and scale, potentially transforming how we understand and treat cancer.
When a patient is suspected of having cancer, a tissue specimen is sometimes removed, stained, affixed to a glass slide, and analyzed by a pathologist using a microscope. Pathologists perform several tasks on this tissue like detecting cancerous cells and determining the cancer subtype. Increasingly, these tiny tissue samples are being digitized into enormous whole slide images, detailed enough to be up to 50,000 times larger than a typical photo stored on a mobile phone. The recent success of machine learning models, combined with the increasing availability of these images, has ignited the field of computational pathology, which focuses on the creation and application of machine learning models for tissue analysis and aims to uncover new insights in the fight against cancer.
Until recently, the potential applicability and impact of computational pathology models were limited because these models were diagnostic-specific and typically trained on narrow samples. Consequently, they often lacked sufficient performance for real-world clinical practice, where patient samples represent a broad spectrum of disease characteristics and laboratory preparations. In addition, applications for rare and uncommon cancers struggled to collect adequate sample sizes, which further limited the reach of computational pathology.
The rise of foundation models is introducing a new paradigm in computational pathology. These large neural networks are trained on vast and diverse datasets that do not need to be labeled, making them capable of generalizing to many tasks. They have created new possibilities for learning from large, unlabeled whole slide images. However, the success of foundation models critically depends on the size of both the dataset and model itself.
Advancing pathology foundation models with data scale, model scale, and algorithmic innovation
Microsoft Research, in collaboration with Paige (opens in new tab), a global leader in clinical AI applications for cancer, is advancing the state-of-the-art in computational foundation models. The first contribution of this collaboration is a model named Virchow, and our research about it was recently published in Nature Medicine (opens in new tab). Virchow serves as a significant proof point for foundation models in pathology, as it demonstrates how a single model can be useful in detecting both common and rare cancers, fulfilling the promise of generalizable representations. Following this success, we have developed two second-generation foundation models for computational pathology, called Virchow2 and Virchow2G, (opens in new tab) which benefit from unprecedented scaling of both dataset and model sizes, as shown in Figure 1.
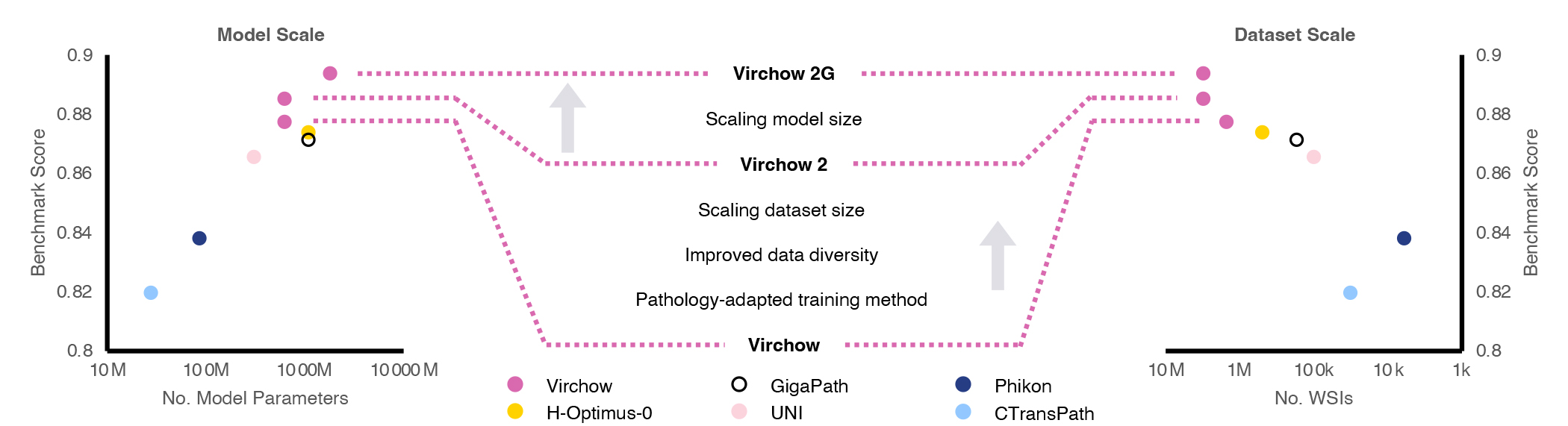
Beyond access to a large dataset and significant computational power, our team demonstrated further innovation by showing how tailoring the algorithms used to train foundation models to the unique aspects of pathology data can also improve performance. These three pillars—data scale, model scale, and algorithmic innovation—are described in a recent technical report.
Virchow foundation models and their performance
Using data from over 3.1 million whole slide images (2.4PB of data) corresponding to over 40 tissues from 225,000 patients in 45 countries, the Virchow2 and 2G models are trained on the largest known digital pathology dataset. Virchow2 matches the model size of the first generation of Virchow with 632 million parameters, while Virchow2G scales model size to 1.85 billion parameters, making it the largest pathology model.
In the report, we evaluate the performance of these foundation models on twelve tasks, aiming to capture the breadth of application areas for computational pathology. Early results suggest that Virchow2 and Virchow2G are better at identifying tiny details in cell shapes and structures, as illustrated in Figure 2. They perform well in tasks like detecting cell division and predicting gene activity. These tasks likely benefit from quantification of nuanced features, such as the shape and orientation of the cell nucleus. We are currently working to expand the number of evaluation tasks to include even more capabilities.

Looking forward
Foundation models in healthcare and life sciences have the potential to significantly benefit society. Our collaboration on the Virchow models has laid the groundwork, and we aim to continue working on these models to provide them with more capabilities. At Microsoft Research Health Futures, we believe that further research and development could lead to new applications for routine imaging, such as biomarker prediction, with the goal of more effective and timely cancer treatments.
Paige has released Virchow2 on Hugging Face (opens in new tab), and we invite the research community to explore the new insights that computational pathology models can reveal. Note that Virchow2 and Virchow2G are research models and are not intended to make diagnosis or treatment decisions.





