In many real-life scenarios, obtaining information is costly, and getting fully observed data is almost impossible. For example, in the recruiting world, obtaining relevant information (in other words, a feature value) for a company could mean performing time-consuming interviews. The same applies to many other scenarios, such as in education and the medical field, where each feature value is an often more complex answer to a question. Unfortunately, AI-aided decision making usually requires large amounts of data. Microsoft researchers, in project Minimum Data AI, aim to investigate how to best utilize AI algorithms to aid decision making while simultaneously minimizing data requirements.
In our paper accepted at the thirty-third Conference on Neural Information Processing Systems (NeurIPS 2019), titled “Icebreaker: Element-wise efficient information acquisition with a Bayesian Deep Latent Gaussian Model,” Microsoft researchers, along with researchers in the Department of Engineering at University of Cambridge, tackle the challenge of deploying machine learning models when very little or no training data is initially available and also when acquiring each feature element of data is associated with costs. We call this challenge the ice-start problem. To solve this problem, we propose the Icebreaker solution, a novel deep generative model that minimizes the amount and cost of data required to train a machine learning model. This work can be combined with our previous work, EDDI, which performs efficient information acquisition in the test time.

Spotlight: Blog post
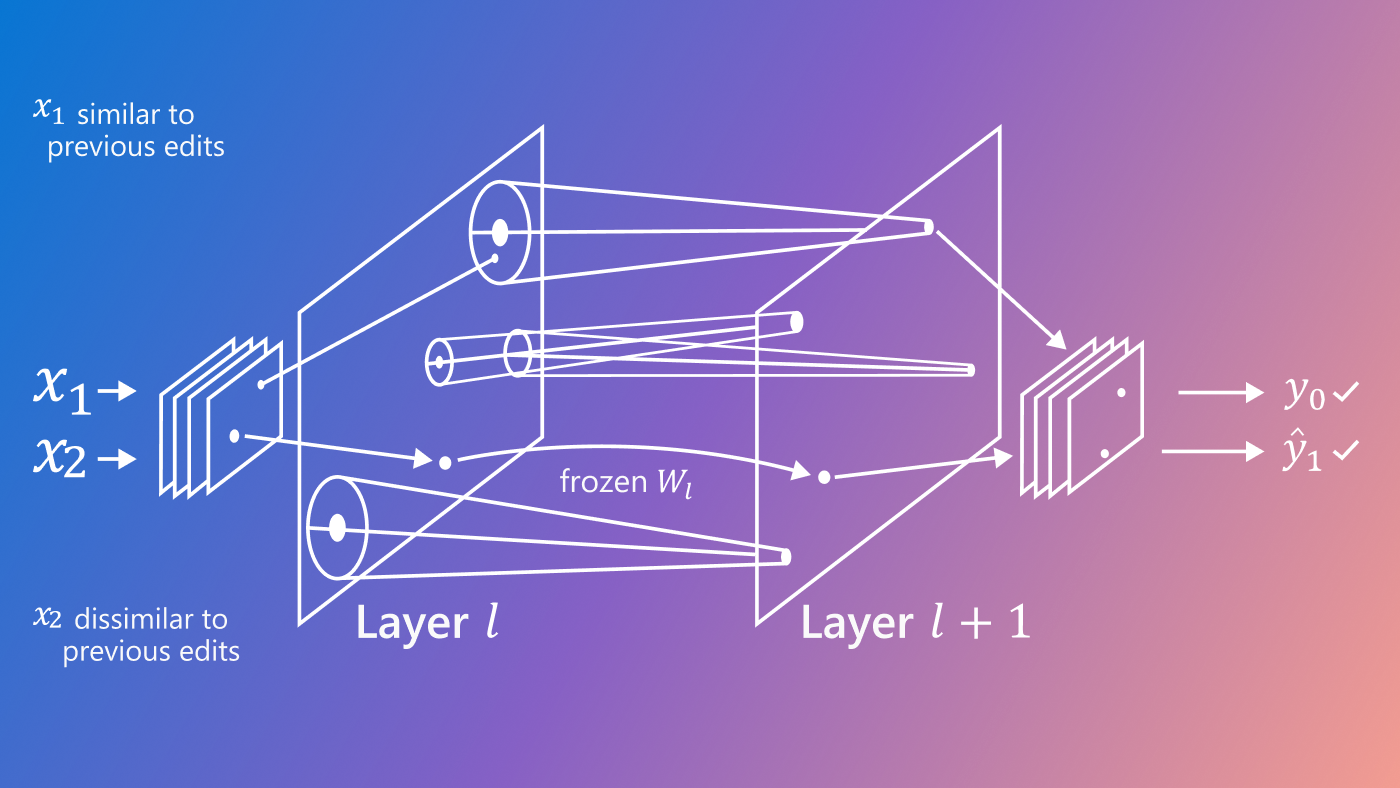
Figure 1: Our model employs two components. The first component is a deep generative model (PA-BELGAM), shown in the top half of the model above, which features a novel inference algorithm that can explicitly quantify epistemic uncertainty. The second component is a set of new element-wise training data selection objectives for data acquisition, shown in the bottom half of the model.
Partial Amortized Bayesian Deep Latent Gaussian Model (PA-BELGAM)
To enable element-wise training data selection for a flexible model with explicit parameter uncertainty modelling, our work consists of two components: the first part is a deep generative model called the Partial Amortized Bayesian Deep Latent Gaussian Model (PA-BELGAM) with a novel inference algorithm, which can explicitly quantify epistemic uncertainty and can be trained with any volume of partially observed data. The second part is a set of new element-wise training data selection objectives for element-wise training data acquisition.

Figure 2: PA-BELGAM, a deep latent gaussian model with explicit parameter uncertainty estimation, is also able to handle any subset of missing feature values.
The PA-BELGAM model is based on the variational autoencoder model. The key differences are the ability to handle missing elements of the data and the Bayesian treatment of the decoder weights θ. Instead of using a standard deep neural network as the decoder to map data from a latent representation, we use a Bayesian neural network, and we put a prior distribution over the decoder weights θ.
The design of PA-BELGAM leads to the new challenge of having to approximate the intractable posterior of the decoder weights and the latent variables in an efficient and accurate way. Our solution is to combine the efficiency of amortized inference with the accuracy of sampling methods. In general, the inference consists of an encoder parameter update using amortized inference followed by a decoder sampling using Stochastic Gradient MCMC.
To enable the active element-wise training data selection, we then designed element-wise data selection objectives for different applications, considering both unsupervised settings (such as matrix imputation tasks, which can be used as a recommender), and supervised settings (such as classification or regression tasks). For imputation tasks, the objective aims to select the feature element that maximizes the reduction of the model parameter uncertainty. For the prediction tasks, the objective not only handles traditional one-step prediction but also considers active sequential prediction (such as EDDI). We designed the objective to both reduce the model parameter uncertainty and maximize the predictive power.
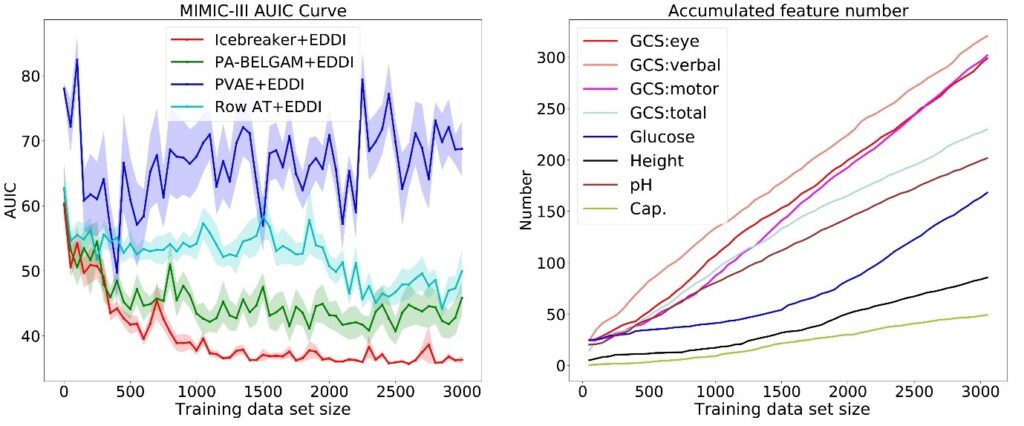
Figure 3: Results showing use of Icebreaker for training feature element acquisition on the MIMIC data set.
As an example, we show our method using the largest publicly available medical dataset called MIMIC (note: this dataset does not use protected health information). We use Icebreaker for training feature element acquisition and combine it with our prior work on test time information acquisition and prediction called EDDI. In Figure 3, the graph on the left shows that our proposed method performs better than several baselines, achieving better test accuracy with less training data. The graph on the right in Figure 3 shows the number of data points for eight features as the total size of our data set grows. The non-linear growth for some of the features (for example, Glucose) indicates that our method is able to learn to select different features at different stages of the training procedures. For more experiments, please check out our NeurIPS paper.
We have released our code online on GitHub. We encourage you to experiment with the whole framework—train a PA-BELGAM model with element-wise training data selection or simply try out PA-BELGAM as an alternative to variational autoencoders for your application. To learn more about the Minimum Data AI project, please visit our project page.
If you will be attending NeurIPS 2019 and are interested in learning about this work or the Minimum Data AI project, come find us at our poster session on Wednesday, December 11th, from 5:00 PM – 7:00 PM PST in East Exhibition Hall B and C. We will be there and will be happy to speak with you more about our research. Feel free to stop by and meet us at the Microsoft booth as well!