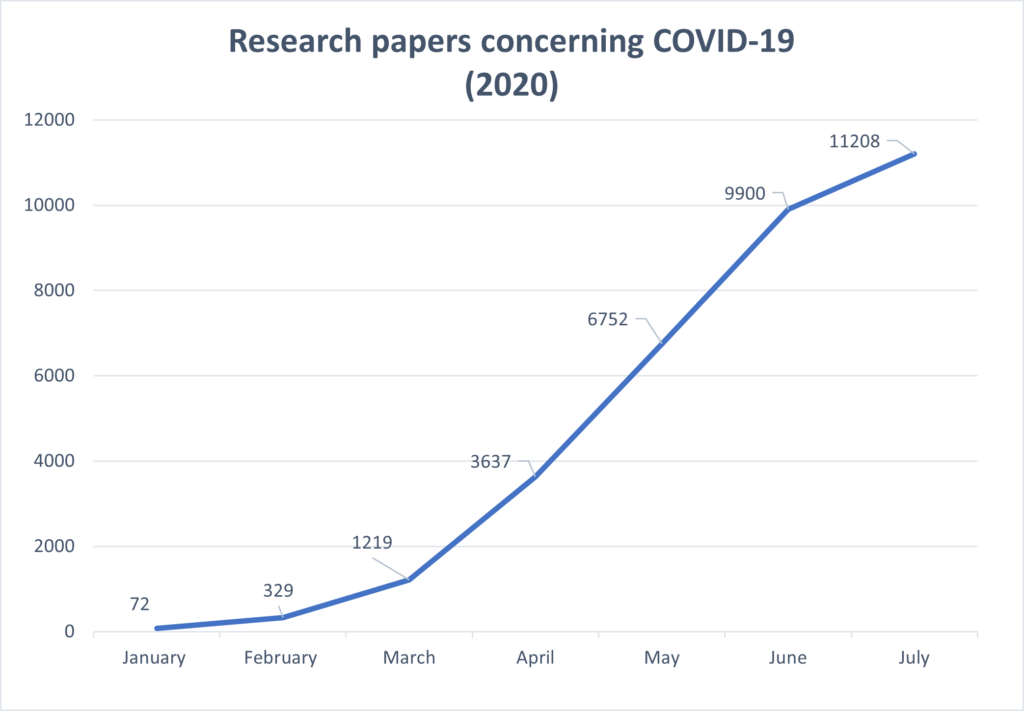
COVID-19 highlights a perennial problem facing scientists around the globe: how do we stay up to date with the cutting edge of scientific knowledge? In just a few months since the pandemic emerged, tens of thousands of research papers have been published concerning COVID-19 and the SARS-CoV-2 virus. This explosive growth sparks the creation of the COVID-19 Open Research Dataset (CORD-19) to facilitate research and discovery. However, a pandemic is just one salient example of a prevailing challenge to this community. PubMed, the standard repository for biomedical research articles, adds 4,000 new papers every day and over a million every year.
It is impossible to keep track of such rapid progress by manual efforts alone. In the era of big data and precision medicine, the urgency has never been higher to advance natural language processing (NLP) methods that can help scientists stay versed in the deluge of information. NLP can help researchers quickly identify and cross-reference important findings in papers that are both directly and tangentially related to their own research at a large scale—instead of researchers having to sift through papers manually for relevant findings or recall them from memory.
In this blog post, we present our recent advances in pretraining neural language models for biomedical NLP. We question the prevailing assumption that pretraining on general-domain text is necessary and useful for specialized domains such as biomedicine. Instead, we show that biomedical text is very different from newswires and web text. By pretraining solely on biomedical text from scratch, our PubMedBERT model outperforms all prior language models and obtains new state-of-the-art results in a wide range of biomedical applications. To help accelerate progress in this vitally important area, we have created a comprehensive benchmark and released the first leaderboard for biomedical NLP. Our findings might also be potentially applicable to other high-value domains, such as finance and law.
A new paradigm for building neural language models in biomedicine and specialized domains
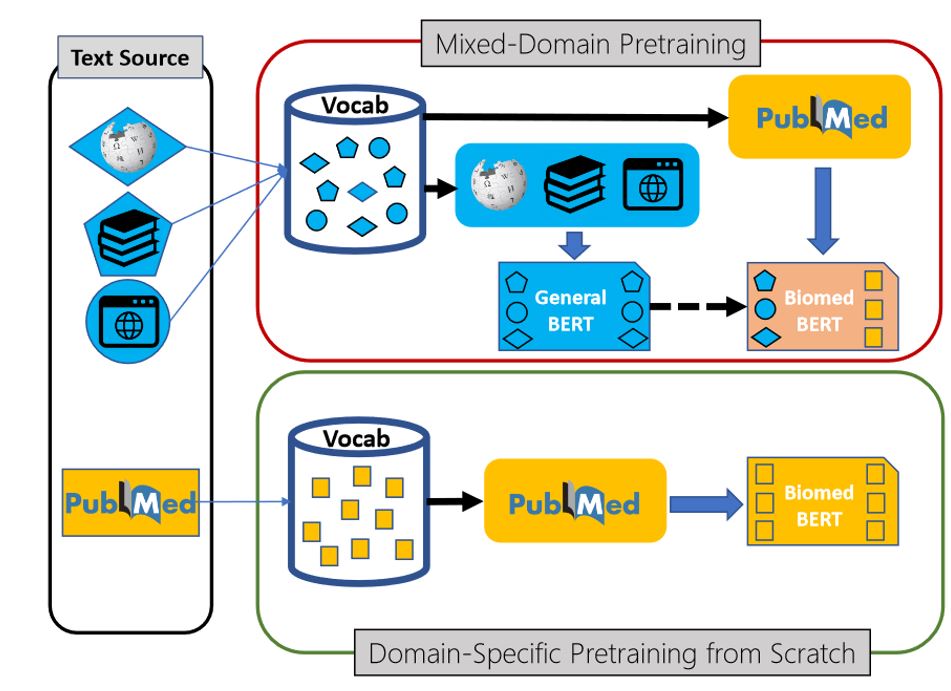
Pretrained neural language models are the underpinning of state-of-the-art NLP methods. Pretraining works by masking some words from text and training a language model to predict them from the rest. Then, the pre-trained model can be fine-tuned for various downstream tasks using task-specific training data. As in mainstream NLP, prior work on pretraining is largely concerned about newswires and the Web. For applications in such general domains, the topic is not known a priori, it is thus advantageous to train a broad-coverage model using as much text as one could gather.
Microsoft research podcast
Collaborators: Silica in space with Richard Black and Dexter Greene
College freshman Dexter Greene and Microsoft research manager Richard Black discuss how technology that stores data in glass is supporting students as they expand earlier efforts to communicate what it means to be human to extraterrestrials.
For specialized domains like biomedicine, which has abundant text that is drastically different from general-domain corpora, this rationale no longer applies. PubMed contains over 30 million abstracts, and PubMed Central (PMC) contains millions of full-text articles. Still, the prevailing assumption is that out-domain text, in this case text not related to biomedicine, can be helpful, so prior work typically adopts a mixed-domain approach by starting from a general-domain language model.
| Biomedical Term | Category | BERT | SciBERT | PubMedBERT (Ours) |
| diabetes | disease | X | X | X |
| leukemia | disease | X | X | X |
| lithium | drug | X | X | X |
| insulin | drug | X | X | X |
| DNA | gene | X | X | X |
| promoter | gene | X | X | X |
| hypertension | disease | X | X | |
| nephropathy | disease | X | X | |
| lymphoma | disease | X | X | |
| lidocaine | drug | X | X | |
| oropharyngeal | organ | X | ||
| cardiomyocyte | cell | X | ||
| chloramphenicol | drug | X | ||
| RecA | gene | X | ||
| acetyltransferase | gene | X | ||
| clonidine | drug | X | ||
| naloxone | drug | X |
We challenge this assumption and propose a new paradigm that pretrains entirely on in-domain text from scratch for a specialized domain. We observe that biomedical text is very different from general-domain text. As shown in the above figure, the standard BERT model pretrained on general-domain text only covers the most frequent biomedical terms. Others will be shattered to non-sensical sub-words. For example, lymphoma is represented as l, ##ym, ##ph, or ##oma. Acetyltransferase is reduced to ace, ##ty, ##lt, ##ran, ##sf, ##eras, or ##e. By contrast, our PubMedBERT treats biomedical terms as “first-class citizens” and avoids diverting precious modeling and compute bandwidth to irrelevant out-domain text.
Creating a comprehensive benchmark and leaderboard to accelerate progress in Biomedical NLP
Comprehensive benchmarks and leaderboards, such as GLUE (opens in new tab), have greatly accelerated progress in general NLP. For biomedicine, however, such benchmarks and leaderboards are ostensibly absent. Prior work tends to use different tasks and datasets for downstream evaluation, which makes it hard to assess the true impact of biomedical pretraining strategies.
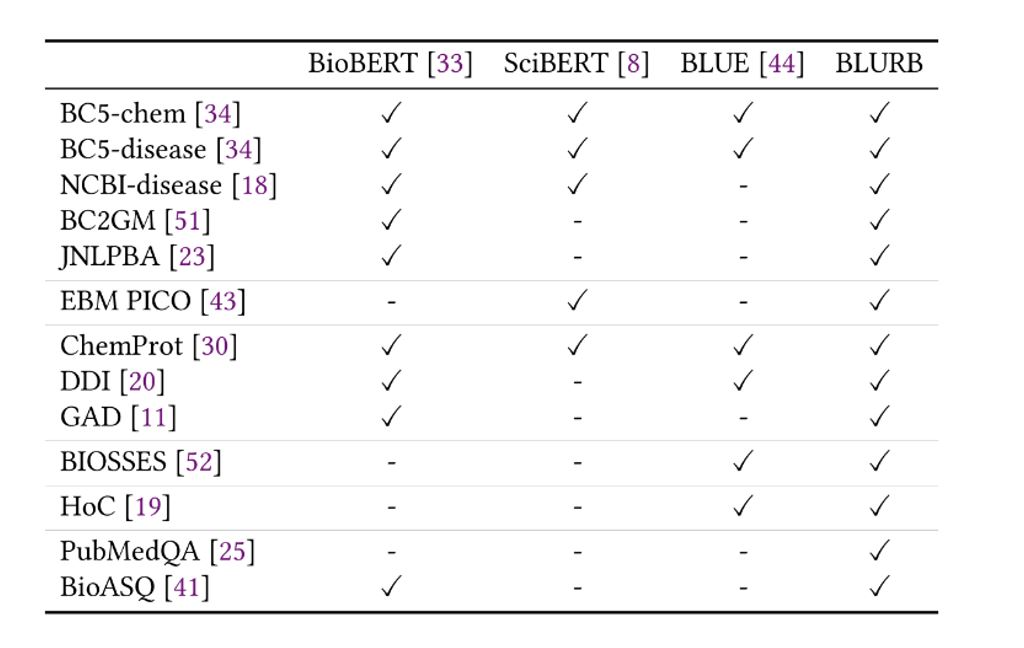
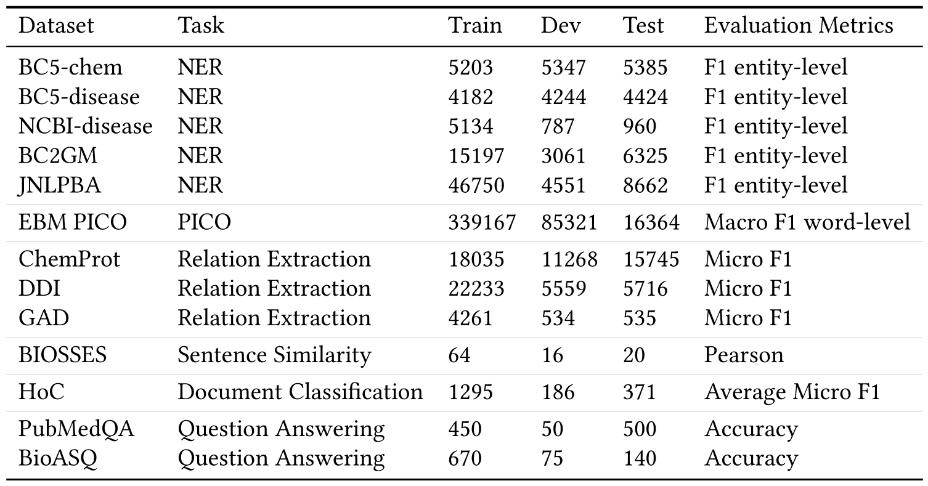
To address this problem, we have created the Biomedical Language Understanding and Reasoning Benchmark (opens in new tab)(BLURB) for PubMed-based biomedical NLP applications. BLURB consists of 13 publicly available datasets in six diverse tasks including: named entity recognition, evidence-based medical information extraction, relation extraction, sentence similarity, document classification, and question answering (see Table 3). To avoid placing undue emphasis on tasks with many available datasets, such as named entity recognition (NER), BLURB reports the macro average across all tasks as the main score. We also have created a leaderboard to track progress by the community. The BLURB leaderboard is model agnostic. Any system capable of producing the test predictions using the same training and development data can participate. The main goal of BLURB is to lower the entry barrier into biomedical NLP and help accelerate progress in this vitally important area for societal and human impact.
PubMedBERT: outperforming all prior language models and attaining state-of-the-art biomedical NLP results
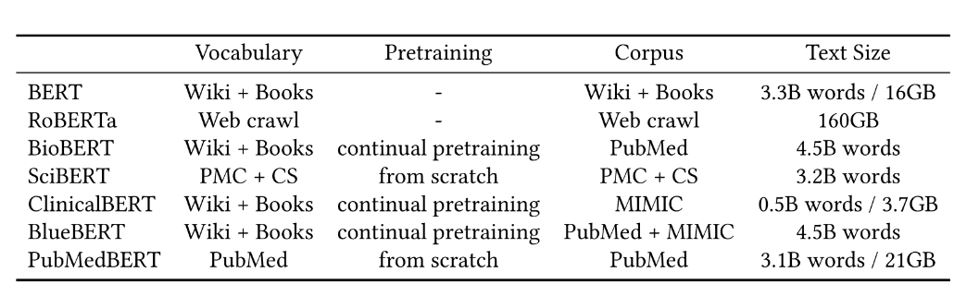
We pretrain our PubMedBERT model on biomedical text from scratch. The pretraining corpus comprises 14 million PubMed abstracts with 3 billion words (21 GB), after filtering empty or short abstracts. To enable fair comparison, we use the same amount of compute as in prior biomedical pretraining efforts. We also pretrain another version of PubMedBERT by adding full-text articles from PubMed Central, with the pretraining corpus increased substantially to 16.8 billion words (107 GB).
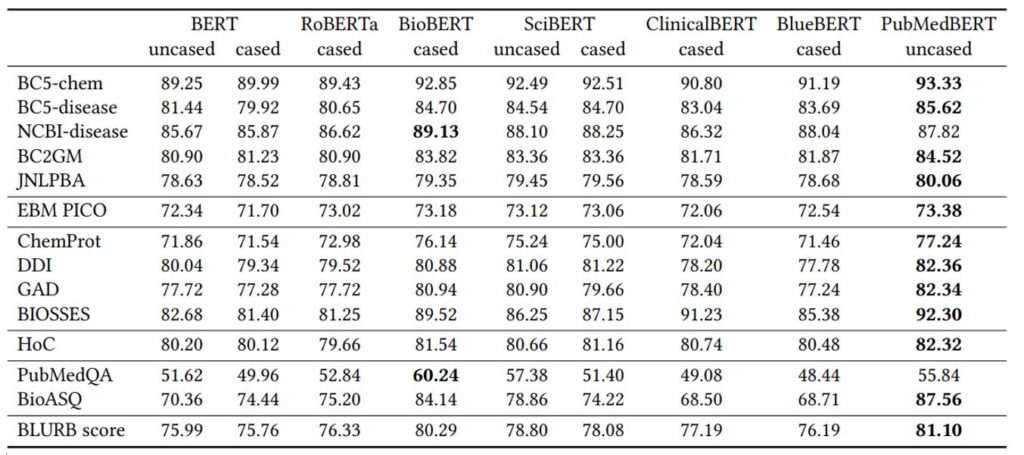
As Table 5 shows, PubMedBERT consistently outperforms all prior language models across biomedical NLP applications, often by a significant margin. The gains are most substantial against general-domain models. Most notably, while RoBERTa uses the largest pretraining corpus, its performance on biomedical NLP tasks is among the worst, similar to the original BERT model. Models using biomedical text in pretraining generally perform better. However, mixing out-domain text in pretraining generally leads to worse performance. In particular, even though clinical notes are more relevant to the biomedical domain than general-domain text, adding them does not confer any advantage, as evident by the results of ClinicalBERT and BlueBERT. Not surprisingly, BioBERT is the closest to PubMedBERT, as it also uses PubMed text for pretraining. However, by conducting domain-specific pretraining from scratch, PubMedBERT is able to obtain consistent gains over BioBERT in most tasks.
Analysis of pretraining and task-specific fine-tuning strategies
In addition to establishing the superiority of domain-specific pretraining, we have also conducted thorough analyses of the myriad choices in pretraining and task-specific fine-tuning, with several interesting findings:
Pretraining on full-text articles generally leads to a slight degradation in performance compared with pretraining using only abstracts. We hypothesize that the reason is twofold. First, full text is generally noisier than abstracts. As existing biomedical NLP tasks are mostly based on abstracts, full text may be even slightly out-domain compared to abstracts. Moreover, even if full text can be helpful, their inclusion requires additional pretraining cycles to make use of extra information. Indeed, by extending pretraining for 60% longer, the overall results are slightly better than that of the standard PubMedBERT using only abstracts.
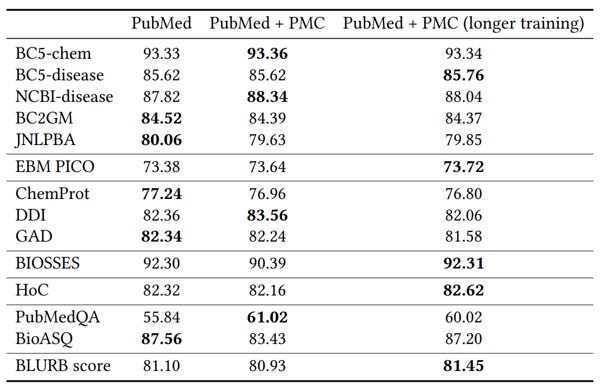
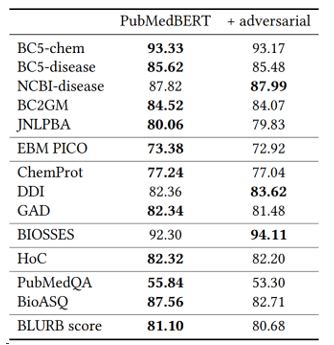
PubMed''). The second one corresponds to PubMedBERT trained using both PubMed abstracts and PubMed Central full text (PubMed+PMC”). The last one corresponds to PubMedBERT trained using both PubMed abstracts and PubMed Central full text, for 60% longer (“PubMed+PMC (longer training)”).Adversarial pretraining (opens in new tab) has been shown to be highly effective in boosting performance in general-domain applications. Surprisingly, it generally leads to a slight degradation in PubMedBERT performance (see Table 7). We hypothesize that the reason may be similar to what we observe in pretraining with full texts. Namely, adversarial training is more useful if the pretraining corpus is more diverse and relatively out-domain compared to the application tasks.
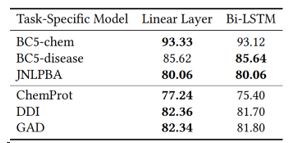
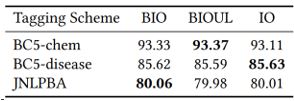
Some common practices in named entity recognition and relation extraction may no longer be necessarily with the use of neural language models. Specifically, with the use of self-attention mechanism, the utility in explicit sequential modeling becomes questionable. In ablation studies, we find that a linear layer performs comparable or better than sequential modeling methods such as bi-directional LSTMs. For named entity recognition, the tagging scheme that simply differentiates between inside and outside of entity mentions suffices.
A pretraining method for specialized domains that complements generic language models
To reiterate, we propose a new paradigm for domain-specific pretraining by learning neural language models from scratch entirely within a specialized domain. We show that for high-volume, high-value domains such as biomedicine, such a strategy outperforms all prior language models and obtains state-of-the-art results across the board in biomedical NLP applications.
This strategy is not in conflict with pretraining a generic all-encompassing language model. Indeed, the two complement each other and are applicable in different situations. In many open-domain applications (such as search, ads, productivity suites, and others), one can’t anticipate the domain for the application instances. Therefore, a general language model accounting for as many domains as possible is mandatory. On the other hand, for applications in a specialized domain with abundant text, domain-specific pretraining is superior and desirable. In biomedicine, prominent examples include extracting PubMed-scale knowledge graphs and compiling population-level real-world evidence from electronic medical records.
This work builds on past advances in biomedical NLP (opens in new tab) and deep learning (opens in new tab) at Microsoft Research. We are sharing our comprehensive benchmark, BLURB, and the first leaderboard for biomedical NLP at http://aka.ms/blurb (opens in new tab). We hope these resources will help lower the barrier for entry and spur progress in this vitally important area. We have also released the state-of-the-art PubMedBERT and task-specific models at the BLURB website, which can be found under the “Model” tab there. We encourage researchers to participate in the leaderboard, and we hope that you will download and apply PubMedBERT and task-specific models to your own work.





