
Editor’s note, Sept. 28, 2023 – The founding collaborators list was updated to correct omissions and the scientific foundation model graph was updated to correct information.
Introduction
In the next decade, deep learning may revolutionize the natural sciences, enhancing our capacity to model and predict natural occurrences. This could herald a new era of scientific exploration, bringing significant advancements across sectors from drug development to renewable energy. In line with Microsoft’s mission to empower every person and every organization on the planet to achieve more, the DeepSpeed (opens in new tab) team at Microsoft is responding to this opportunity by launching a new initiative called DeepSpeed4Science (opens in new tab), aiming to build unique capabilities through AI system technology innovations to help domain experts to unlock today’s biggest science mysteries.
The DeepSpeed (opens in new tab) system is an industry leading open-source AI system framework, developed by Microsoft, that enables unprecedented scale and speed for deep learning training and inference on a wide range of AI hardware. Figure 1 demonstrates our basic approach to this new initiative. By leveraging DeepSpeed’s current technology pillars (training, inference and compression) as base technology enablers, DeepSpeed4Science will create a new set of AI system technologies tailored for accelerating scientific discoveries by addressing their unique complexity beyond the common technical approaches used for accelerating generic large language models (LLMs). We work closely with internal and external teams who own AI-driven science models that represent key science missions, to identify and address general domain-specific AI system challenges. This includes climate science, drug design, biological understanding, molecular dynamics simulation, cancer diagnosis and surveillance, catalyst/material discovery, and other domains.
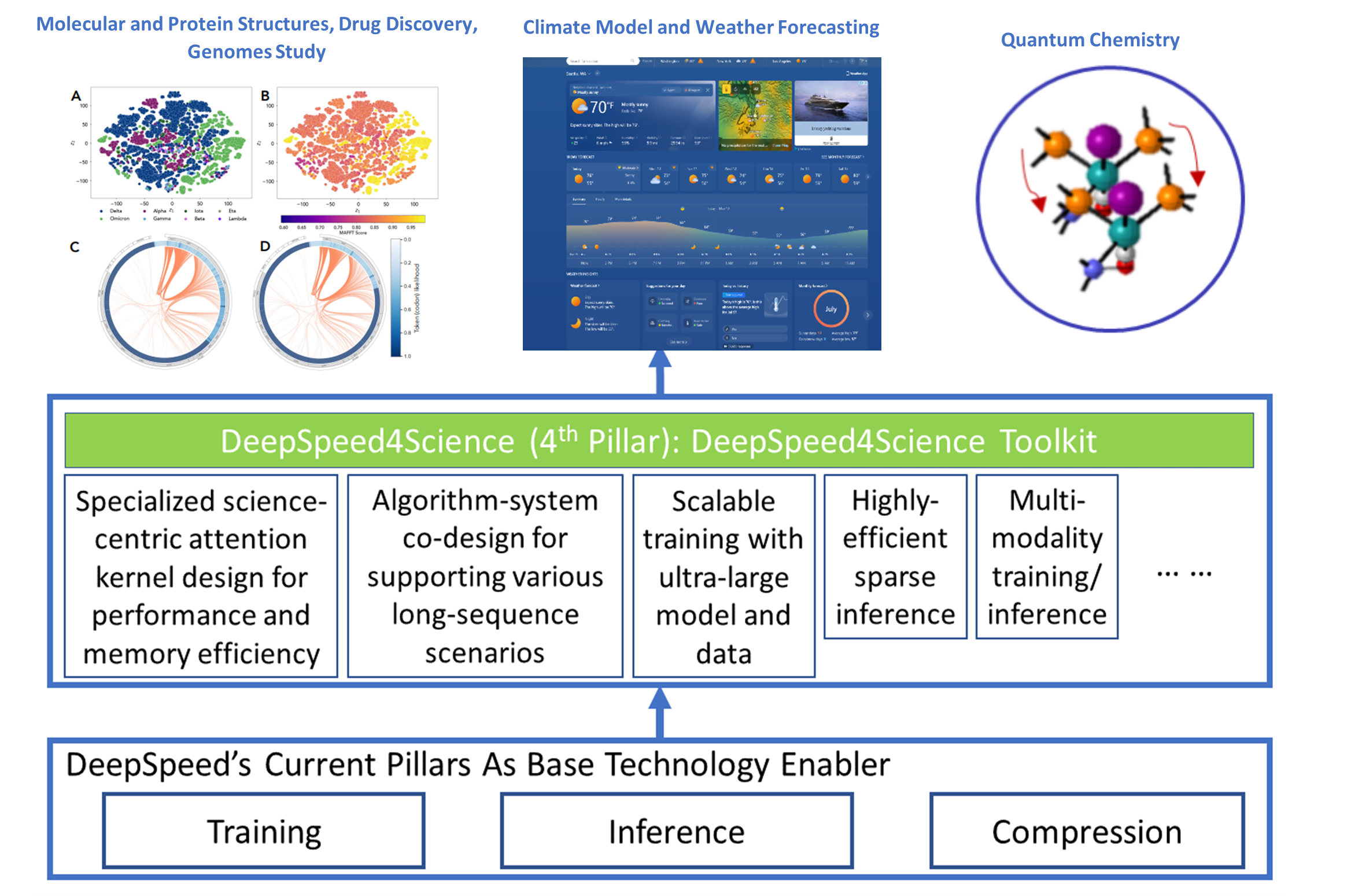
Our long-term vision is to develop DeepSpeed4Science into a new platform and a unified repository for sharing advanced AI system technologies that support scientific discoveries. DeepSpeed4Science is designed to be inclusive, echoing Microsoft’s AI for Good commitment. That is reflected in the initiative’s support for a diverse group of signature science models, representing some of the most critical AI for science investments. In this blog, we showcase how DeepSpeed4Science helps address two of their critical system challenges in structural biology research: (1) eliminating memory explosion problems for scaling Evoformer-centric protein-structure prediction models, and (2) enabling very-long sequence support for better understanding the evolutionary landscape of pandemic-causing viruses.
Our launch and key collaborators
The new system technologies enabled by DeepSpeed4Science can empower AI-driven scientific discoveries using signature models that represent a wide spectrum of efforts pushing the boundaries of science. Currently, DeepSpeed4Science is honored to support several key science models from Microsoft Research AI4Science (opens in new tab), Microsoft WebXT/Bing (opens in new tab) and U.S. DoE National Labs (opens in new tab).
Current Microsoft internal partnerships
Scientific Foundation Model (SFM), Microsoft Research AI4Science

Scientific foundation model (SFM) aims to create a unified large-scale foundation model to empower natural scientific discovery by supporting diverse inputs, multiple scientific domains (e.g., drugs, materials, biology, health, etc.) and computational tasks. The DeepSpeed4Science partnership will provide new training and inference technologies to empower the SFM team’s continuous research on projects like Microsoft’s new generative AI methods, such as Distributional Graphormer.
ClimaX, MSR AI4Science
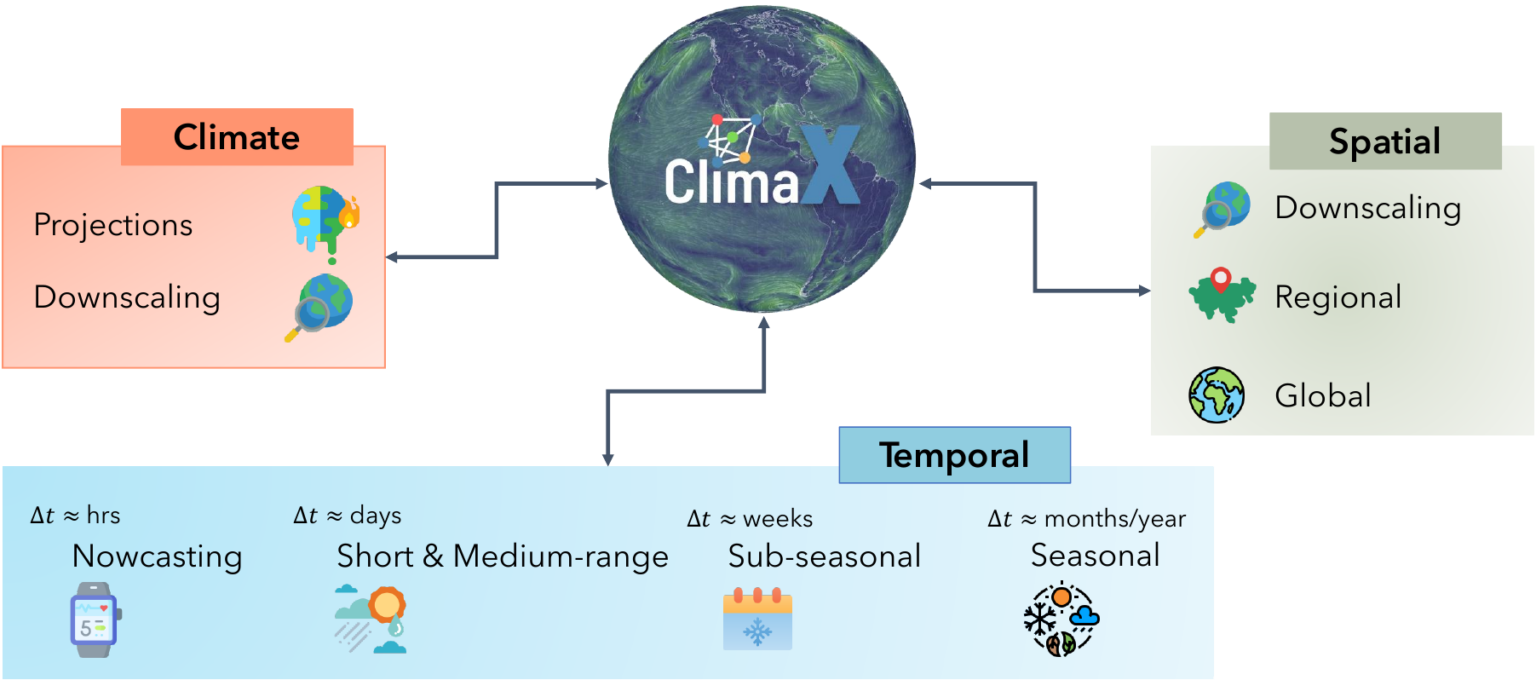
Our changing climate is producing more frequent extreme weather events. To mitigate the negative effects, it is increasingly important to predict where these events will occur. ClimaX is the first foundation model designed to perform a wide variety of weather and climate modeling tasks. It can absorb many different datasets with different variables and resolutions, potentially improving weather forecasting. DeepSpeed4Science is creating new system supports and acceleration strategies for ClimaX for efficiently pretraining/finetuning bigger foundation models while handling very large high-resolution image data (e.g., tens to hundreds of petabytes) with long sequences.
AI Powered Ab Initio Molecular Dynamics (AI2MD), MSR AI4Science
This project simulates the dynamics of large (million-atom) molecular systems with near ab initio accuracy using AI-powered force field models while maintaining the efficiency and scalability of classical molecular dynamics. The simulations are efficient enough to generate trajectories long enough to observe chemically significant events. Typically, millions or even billions of inference steps are required for this process. This poses a significant challenge in optimizing the inference speed of graph neural network (GNN)+ LLM models, for which DeepSpeed4Science will provide new acceleration strategies.
Weather from Microsoft Start, Microsoft WebXT/Bing
Weather from Microsoft Start (opens in new tab) provides precise weather information to help users make better decisions for their lifestyles, health, jobs and activities (opens in new tab) – including accurate 10-day global weather forecasts updated multiple times every hour. Previously, Weather from Microsoft Start benefited from DeepSpeed technologies to accelerate their multi-GPU training environments. Currently, DeepSpeed4Science is working with the WebXT weather team to further enhance Microsoft Weather services with cutting-edge features and improvements.
Current external collaborators
DeepSpeed4Science’s journey started with two pioneering LLM-based AI models for structural biology research: OpenFold (opens in new tab) from Columbia University, an open-sourced high-fidelity protein structure prediction model; and GenSLMs (opens in new tab) from Argonne National Laboratory (opens in new tab), an award-winning genome-scale language model (opens in new tab) for learning the evolutionary landscape of SARS-CoV-2 (COVID-19) genomes. As the featured showcases for this release, they represent two common AI system challenges facing today’s AI-driven structural biology research. We will discuss how DeepSpeed4Science empowered their scientific discovery in the next section.
Additionally, DeepSpeed4Science has recently expanded its scope to support a more diverse range of science models. For example, in our work with Argonne on training a trillion-parameter science model on Aurora Exascale system (opens in new tab), DeepSpeed4Science technologies will help them reach the performance requirements and scalability needed for this critical mission. Furthermore, by collaborating with Oak Ridge National Lab (opens in new tab) and National Cancer Institute (NCI) (opens in new tab) on cancer surveillance, DeepSpeed4Science will help enable high-fidelity extraction and classification of information from unstructured clinical texts for the MOSSAIC project (opens in new tab). DeepSpeed4Science technologies will also be adopted by Brookhaven National Laboratory (opens in new tab) to support development of a large digital twin model for clean energy research by using LLMs to produce more realistic simulation data. You can find more detailed information about our external colleagues and their science missions at DeepSpeed4Science (opens in new tab).
Partnership showcases
Showcase (I): DeepSpeed4Science eliminates memory explosion problems for scaling Evoformer-centric structural biology models via DS4Sci_EvoformerAttention
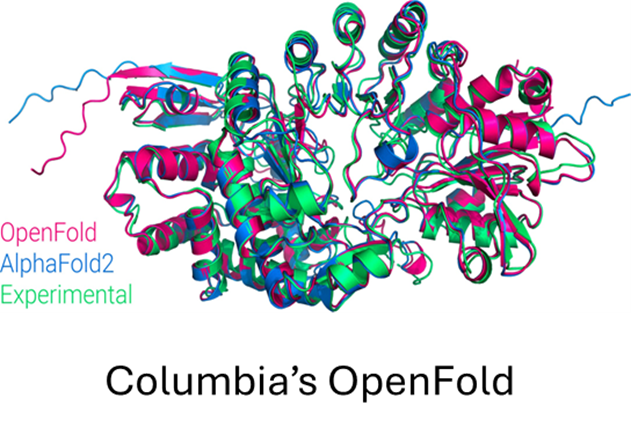
OpenFold (opens in new tab) is a community reproduction of DeepMind’s AlphaFold2 (opens in new tab) that makes it possible to train or finetune AlphaFold2 on new datasets. Researchers have used it to retrain AlphaFold2 from scratch to produce new sets of model parameters, studied the early training phase of AlphaFold2 (Figure 6), and developed new protein folding systems.
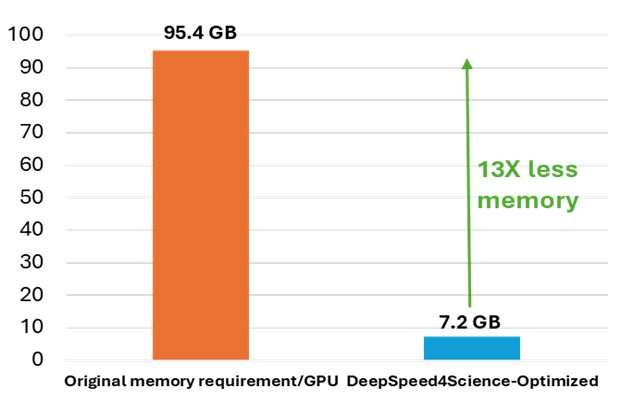
While OpenFold does include performance and memory optimizations using state-of-the-art system technologies, training AlphaFold2 from scratch is still computationally expensive. The model at the current stage is small in absolute terms, with just 93 million parameters, but it contains several custom attention variants that manifest unusually large activations. During the “finetuning” phase of a standard AlphaFold2 training run, the logit tensor produced in just one of these variants–one designed to attend over the deep protein MSAs fed to the model as input–is in excess of 12GB in half precision alone, dwarfing the peak memory requirements of comparably sized language models. Even with techniques like activation checkpointing and DeepSpeed ZeRO optimizations, this memory explosion problem heavily constrains the sequence lengths and MSA depths on which the model can be trained. Furthermore, approximation strategies can significantly affect the model accuracy and convergence, while still resulting in memory explosion, shown as the left bar (orange) in Figure 7.
To address this common system challenge in structural biology research (e.g., protein structure prediction and equilibrium distribution prediction), DeepSpeed4Science is addressing this memory inefficiency problem by designing customized exact attention kernels for the attention variants (i.e., EvoformerAttention), which widely appear in this category of science models. Specifically, a set of highly memory-efficient DS4Sci_EvoformerAttention kernels enabled by sophisticated fusion/tiling strategies and on-the-fly memory reduction methods, are created for the broader community as high-quality machine learning primitives. Incorporated into OpenFold, they provide a substantial speedup during training and dramatically reduce the model’s peak memory requirement for training and inference. This allows OpenFold to be experimented with bigger and more complex models, and longer sequences, and trained on a wider spectrum of hardware. Detailed information about this technology can be found at DeepSpeed4Science (opens in new tab).
Showcase (II): DeepSpeed4Science enables very-long sequence support via both systematic and algorithmic approaches for genome-scale foundation models (e.g., GenSLMs)
GenSLMs (opens in new tab), a 2022 ACM Gordon Bell award (opens in new tab) winning genome-scale language model from Argonne National Lab, can learn the evolutionary landscape of SARS-CoV-2 (COVID-19) genomes by adapting large language models (LLMs) for genomic data. It is designed to transform how new and emergent variants of pandemic-causing viruses, especially SARS-CoV-2, are identified and classified. GenSLM represents one of the first whole genome-scale foundation models which can generalize to other prediction tasks. A good understanding of the latent space can help GenSLMs tackle new domains beyond just viral sequences and expand their ability to model bacterial pathogens and even eukaryotic organisms, e.g., to understand things such as function, pathway membership, and evolutionary relationships. To achieve this scientific goal, GenSLMs and similar models require very long sequence support for both training and inference that is beyond generic LLMs’ long-sequence strategies like FlashAttention (opens in new tab). Through DeepSpeed4Science’s new designs, scientists can now build and train models with significantly longer context windows, allowing them to explore relationships that were previously inaccessible.
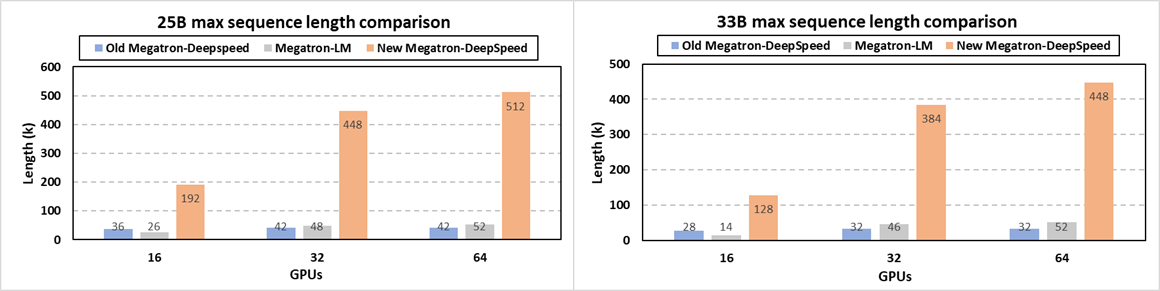
Specifically, at system level, we release the newest Megatron-DeepSpeed (opens in new tab) framework for very-long sequence support along with other new optimizations (opens in new tab). Scientists can now train their large science models like GenSLMs with much longer sequences via a synergetic combination of our newly added memory optimization techniques on attention mask and position embedding, tensor parallelism, pipeline parallelism, sequence parallelism, ZeRO-style data parallelism and model state offloading. Figure 9 demonstrates that our new release enables the longest sequence length for GenSLMs’ 25B and 33B models by up to 12X and 14X, respectively, over the previous Megatron-DeepSpeed. In terms of supported sequence lengths, this new framework also significantly outperforms NVIDIA’s Megatron-LM by up to 9.8X and 9.1X for the 25B and 33B models, respectively. For example, GenSLMs’ 25B model can now be trained with a 512K sequence of nucleotides, compared to the Argonne team’s original 42K sequence length on 64 GPUs. This drastically improves model quality and scientific discovery scope with no accuracy loss. Additional support for domain scientists who prefer algorithmic strategies like relative position embedding techniques is also integrated in this new release (opens in new tab).
Summary and roadmap
We are very proud and excited to announce the DeepSpeed4Science initiative along with several R&D highlights and achievements. Starting today, we will host our new initiative at DeepSpeed4Science (opens in new tab), including information about our external colleagues, and current and future DeepSpeed4Science technology releases. One of our high-level goals is to generalize AI system technologies that broadly address the major system pain points for large-scale scientific discoveries. We hope scientists around the world will enjoy the new capabilities unlocked by DeepSpeed4Science through open-sourced software. We are looking forward to better understanding the AI system design challenges that block your discovery progress. We sincerely welcome your participation to help us build a promising AI4Science future. Please email us at [email protected] (opens in new tab). We encourage you to report issues, contribute PRs, and join discussions on our DeepSpeed GitHub (opens in new tab) page.
Acknowledgements
Core DeepSpeed4Science Team:
Shuaiwen Leon Song (DeepSpeed4Science lead), Minjia Zhang, Conglong Li, Shiyang Chen, Chengming Zhang, Xiaoxia (Shirley) Wu, Masahiro Tanaka, Martin Cai, Adam Graham, Charlie Zhou, Yuxiong He (DeepSpeed team lead)
Our Founding Collaborators (in alphabetical order):
Argonne National Lab team: Rick Stevens, Cristina Negri, Rao Kotamarthi, Venkatram Vishwanath, Arvind Ramanathan, Sam Foreman, Kyle Hippe, Troy Arcomano, Romit Maulik, Maxim Zvyagin, Alexander Brace, Bin Zhang, Cindy Orozco Bohorquez, Austin Clyde, Bharat Kale, Danilo Perez-Rivera, Heng Ma, Carla M. Mann, Michael Irvin, J. Gregory Pauloski, Logan Ward, Valerie Hayot, Murali Emani, Zhen Xie, Diangen Lin, Maulik Shukla, Thomas Gibbs, Ian Foster, James J. Davis, Michael E. Papka, Thomas Brettin
AMD: Ashwin Aji, Angela Dalton, Michael Schulte, Karl Schulz
Brookhaven National Lab team: Adolfy Hoisie, Shinjae Yoo, Yihui Ren.
Columbia University OpenFold team: Mohammed AlQuraishi, Gustaf Ahdritz
Microsoft Research AI4Science team: Christopher Bishop, Bonnie Kruft, Max Welling, Tie-Yan Liu, Christian Bodnar, Johannes Brandsetter, Wessel Bruinsma, Chan Cao, Yuan-Jyue Chen, Peggy Dai, Patrick Garvan, Liang He, Elizabeth Heider, PiPi Hu, Peiran Jin, Fusong Ju, Yatao Li, Chang Liu, Renqian Luo, Qi Meng, Frank Noe, Tao Qin, Janwei Zhu, Bin Shao, Yu Shi, Wenlei Shi, Gregor Simm, Megan Stanley, Lixin Sun, Yue Wang, Tong Wang, Zun Wang, Lijun Wu, Yingce Xia, Leo Xia, Shufang Xie, Shuxin Zheng, Jianwei Zhu
NVIDIA: Yuntian Deng, Weili Nie, Josh Romero, Christian Dallago, Arash Vahdat, Chaowei Xiao, Thomas Gibbs, Anima Anandkumar
Oakridge National Lab team: Prasanna Balaprakash, Gina Tourassi, John Gounley, Heidi Hanson, Thomas E Potok, Massimiliano (Max) Lupo Pasini, Kate Evans, Dan Lu, Dalton Lunga, Junqi Yin, Sajal Dash , Feiyi Wang, Mallikarjun Shankar, Isaac Lyngaas, Xiao Wang, Guojing Cong, Pei Zhang, Ming Fan, Siyan Liu
Princeton University: William Tang, Kyle Felker, Alexey Svyatkovskiy (Microsoft liaison)
Rutgers University: Hang Liu
WebXT Weather team: Pete Luferenko, Divya Kumar, Jonathan Weyn, Ruixiong Zhang, Sylwester Klocek, Volodymyr Vragov





